Multicenter analysis of neutrophil extracellular trap dysregulation in adult and pediatric COVID-19
- PMID: 35852866
- PMCID: PMC9534551
- DOI: 10.1172/jci.insight.160332
Multicenter analysis of neutrophil extracellular trap dysregulation in adult and pediatric COVID-19
Abstract
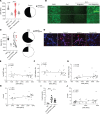
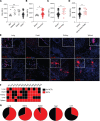
Dysregulation in neutrophil extracellular trap (NET) formation and degradation may play a role in the pathogenesis and severity of COVID-19; however, its role in the pediatric manifestations of this disease, including multisystem inflammatory syndrome in children (MIS-C) and chilblain-like lesions (CLLs), otherwise known as "COVID toes," remains unclear. Studying multinational cohorts, we found that, in CLLs, NETs were significantly increased in serum and skin. There was geographic variability in the prevalence of increased NETs in MIS-C, in association with disease severity. MIS-C and CLL serum samples displayed decreased NET degradation ability, in association with C1q and G-actin or anti-NET antibodies, respectively, but not with genetic variants of DNases. In adult COVID-19, persistent elevations in NETs after disease diagnosis were detected but did not occur in asymptomatic infection. COVID-19-affected adults displayed significant prevalence of impaired NET degradation, in association with anti-DNase1L3, G-actin, and specific disease manifestations, but not with genetic variants of DNases. NETs were detected in many organs of adult patients who died from COVID-19 complications. Infection with the Omicron variant was associated with decreased NET levels when compared with other SARS-CoV-2 strains. These data support a role for NETs in the pathogenesis and severity of COVID-19 in pediatric and adult patients.
Keywords: Infectious disease; Inflammation; Neutrophils.
Conflict of interest statement
Figures


Update of
-
Multicenter analysis of neutrophil extracellular trap dysregulation in adult and pediatric COVID-19.medRxiv [Preprint]. 2022 Mar 3:2022.02.24.22271475. doi: 10.1101/2022.02.24.22271475. medRxiv. 2022. Update in: JCI Insight. 2022 Aug 22;7(16):e160332. doi: 10.1172/jci.insight.160332. PMID: 35262093 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

