Investigating and Modeling the Regulation of Extracellular Antibiotic Resistance Gene Bioavailability by Naturally Occurring Nanoparticles
- PMID: 35853206
- PMCID: PMC9979080
- DOI: 10.1021/acs.est.2c02878
Investigating and Modeling the Regulation of Extracellular Antibiotic Resistance Gene Bioavailability by Naturally Occurring Nanoparticles
Abstract
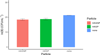
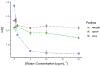
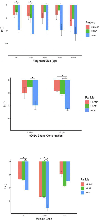
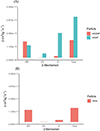
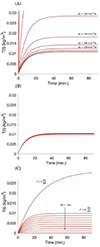
Extracellular antibiotic resistance genes (eARGs) are widespread in the environment and can genetically transform bacteria. This work examined the role of environmentally relevant nanoparticles (NPs) in regulating eARG bioavailability. eARGs extracted from antibiotic-resistant B. subtilis were incubated with nonresistant recipient B. subtilis cells. In the mixture, particle type (either humic acid coated nanoparticles (HASNPs) or their micron-sized counterpart (HASPs)), DNase I concentration, and eARG type were systematically varied. Transformants were counted on selective media. Particles decreased bacterial growth and eARG bioavailability in systems without nuclease. When DNase I was present (≥5 μg/mL), particles increased transformation via chromosomal (but not plasmid-borne) eARGs. HASNPs increased transformation more than HASPs, indicating that the smaller nanoparticle with greater surface area per volume is more effective in increasing eARG bioavailability. These results were also modeled via particle aggregation theory, which represented eARG-bacteria interactions as transport leading to collision, followed by attachment. Using attachment efficiency as a fitting factor, the model predicted transformant concentrations within 35% of experimental data. These results confirm the ability of NPs to increase eARG bioavailability and suggest that particle aggregation theory may be a simplified and suitable framework to broadly predict eARG uptake.
Keywords: antimicrobial resistance; extracellular DNA; horizontal gene transfer; nanoparticles; particle aggregation.
Figures
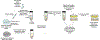

References
-
- He H; Zhou P; Shimabuku K; Fang X; Li S; Lee Y; Dodd M Degradation and Deactivation of Bacterial Antibiotic Resistance Genes During Exposure to Free Chlorine, Monochloramine, Chlorine Dioxide, Ozone, Ultraviolet Light, and Hydroxyl Radical. Environmental Science and Technology 2019, 53, 2013–2026. - PubMed
-
- Allen HK; Donato J; Wang HH; Cloud-Hansen KA; Davies J; Handelsman J Call of the wild: antibiotic resistance genes in natural environments. Nature Reviews Microbiology 2010, 8, 251–259. Magee, J. T.; Pritchard, E.; Fitzgerald, K.; Dunsten, F. D. J.; Howard, A. J. Antibiotic prescribing and antibiotic resistance in community practice: retrospective study. British Medical Journal 1999, 319, 1239–1240. Martínez, J. L. Antibiotics and antibiotic resistance genes in natural environments. Science 2008, 321 (5887), 365–367. Vikesland, P.; Pruden, A.; Alvarez, P.; Aga, D.; Bergman, H.; Li, X.-d.; Manaia, C.; Nambi, I.; Wiggington, K.; Zhang, T.; et al. Toward a Comprehensive Strategy to Mitigate Dissemination of Environmental Sources of Antibiotic Resistance. Environmental Science & Technology: 2017; Vol. 51, pp 13061–13069. - PubMed
-
- Pruden A; Pei R; Storteboom H; Carlson KH Antibiotic resistance genes as emerging contaminants: studies in northern Colorado. Environmental Science and Technology 2006, 40, 7445–7450. - PubMed
-
- Guo XP; Yang Y; Lu DP; Niu ZS; Feng JN; Chen YR; Tou FY; Garner J; Xu J; Liu M; et al. Biofilms as a sink for antibiotic resistance genes (ARGs) in the Yangtze Estuary. Water Research 2017, 129, 277–286. Wang, D. N.; Liu, L.; Qiu, Z. G.; Shen, Z. Q.; Guo, X.; Yang, D.; Li, J.; Liu, W. I.; Jin, M.; Li, J. W. A new adsorption-elution technique for the concentration of aquatic extracellular antibiotic resistance genes from large volumes of water. Water Research 2016, 92, 188–198. Bien, T. L. T.; Thao, N. V.; Kitamura, S. I.; Obayashi, Y.; Suzuki, S. Release and constancy of an antibiotic resistance gene in seawater under grazing stress by ciliates and heterotrophic nanoflagellates. Environmental Microbiology 2017, 32, 174–179. - PubMed
-
- Mao D; Luo Y; Mathieu J; Wang Q; Feng L; Mu Q; Feng C; Alvarez PJJ Persistence of Extracellular DNA in River Sediment Facilitates Antibiotic Resistance Gene Propagation Environmental Science and Technology 2014, 48, 71–78. Yuan, K.; Wang, X.; Chen, X.; Zhao, Z.; Fang, L.; Chen, B.; Jiang, J.; Luan, T.; Chen, B. Occurrence of antibiotic resistance genes in extracellular and intracellular DNA from sediments collected from two types of aquaculture farms. Chemosphere 2019, 234. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

