HGT is widespread in insects and contributes to male courtship in lepidopterans
- PMID: 35853453
- PMCID: PMC9357157
- DOI: 10.1016/j.cell.2022.06.014
HGT is widespread in insects and contributes to male courtship in lepidopterans
Abstract
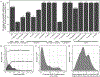
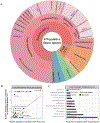
Horizontal gene transfer (HGT) is an important evolutionary force shaping prokaryotic and eukaryotic genomes. HGT-acquired genes have been sporadically reported in insects, a lineage containing >50% of animals. We systematically examined HGT in 218 high-quality genomes of diverse insects and found that they acquired 1,410 genes exhibiting diverse functions, including many not previously reported, via 741 distinct transfers from non-metazoan donors. Lepidopterans had the highest average number of HGT-acquired genes. HGT-acquired genes containing introns exhibited substantially higher expression levels than genes lacking introns, suggesting that intron gains were likely involved in HGT adaptation. Lastly, we used the CRISPR-Cas9 system to edit the prevalent unreported gene LOC105383139, which was transferred into the last common ancestor of moths and butterflies. In diamondback moths, males lacking LOC105383139 courted females significantly less. We conclude that HGT has been a major contributor to insect adaptation.
Keywords: HGT; adaptive evolution; biodiversity; comparative genomics; intron gain; male courtship behavior; symbionts.
Copyright © 2022 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests A.R. is a scientific consultant for LifeMine Therapeutics, Inc.
Figures



References
-
- Anderson DJ (2016). Circuit modules linking internal states and social behaviour in flies and mice. Nat. Rev. Neurosci 17, 692–704. - PubMed
-
- Archibald JM (2015). Endosymbiosis and Eukaryotic Cell Evolution. Curr. Biol 25, R911–R921. - PubMed
-
- Arnold BJ, Huang I-T, and Hanage WP (2022). Horizontal gene transfer and adaptive evolution in bacteria. Nat. Rev. Microbiol 20, 206–218. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

