High-throughput method for the hybridisation-based targeted enrichment of long genomic fragments for PacBio third-generation sequencing
- PMID: 35855323
- PMCID: PMC9278042
- DOI: 10.1093/nargab/lqac051
High-throughput method for the hybridisation-based targeted enrichment of long genomic fragments for PacBio third-generation sequencing
Abstract
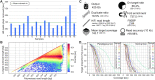
Hybridisation-based targeted enrichment is a widely used and well-established technique in high-throughput second-generation short-read sequencing. Despite the high potential to genetically resolve highly repetitive and variable genomic sequences by, for example PacBio third-generation sequencing, targeted enrichment for long fragments has not yet established the same high-throughput due to currently existing complex workflows and technological dependencies. We here describe a scalable targeted enrichment protocol for fragment sizes of >7 kb. For demonstration purposes we developed a custom blood group panel of challenging loci. Test results achieved > 65% on-target rate, good coverage (142.7×) and sufficient coverage evenness for both non-paralogous and paralogous targets, and sufficient non-duplicate read counts (83.5%) per sample for a highly multiplexed enrichment pool of 16 samples. We genotyped the blood groups of nine patients employing highly accurate phased assemblies at an allelic resolution that match reference blood group allele calls determined by SNP array and NGS genotyping. Seven Genome-in-a-Bottle reference samples achieved high recall (96%) and precision (99%) rates. Mendelian error rates were 0.04% and 0.13% for the included Ashkenazim and Han Chinese trios, respectively. In summary, we provide a protocol and first example for accurate targeted long-read sequencing that can be used in a high-throughput fashion.
© The Author(s) 2022. Published by Oxford University Press on behalf of NAR Genomics and Bioinformatics.
Figures




References
-
- Shendure J., Ji H. Next-generation DNA sequencing. Nat. Biotechnol. 2008; 26:1135–1145. - PubMed
-
- International Human Genome Sequencing Consortium International human genome sequencing consortium. Nature. 2001; 409:860–921. - PubMed
-
- Waterson R., Lindblad-Toh K., Birney E., Rogers J., Abril J. Mouse genome sequencing consortium. Nat. Methods. 2002; 420:61–65.
-
- Salzberg S.L., Yorke J.A. Beware of mis-assembled genomes. Bioinformatics. 2005; 21:4320–4321. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

