Ψ-Footprinting approach for the identification of protein synthesis inhibitor producers
- PMID: 35855324
- PMCID: PMC9290621
- DOI: 10.1093/nargab/lqac055
Ψ-Footprinting approach for the identification of protein synthesis inhibitor producers
Abstract
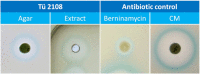
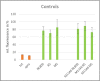
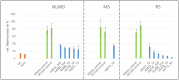
Today, one of the biggest challenges in antibiotic research is a targeted prioritization of natural compound producer strains and an efficient dereplication process to avoid undesired rediscovery of already known substances. Thereby, genome sequence-driven mining strategies are often superior to wet-lab experiments because they are generally faster and less resource-intensive. In the current study, we report on the development of a novel in silico screening approach to evaluate the genetic potential of bacterial strains to produce protein synthesis inhibitors (PSI), which was termed the protein synthesis inhibitor ('psi') target gene footprinting approach = Ψ-footprinting. The strategy is based on the occurrence of protein synthesis associated self-resistance genes in genome sequences of natural compound producers. The screening approach was applied to 406 genome sequences of actinomycetes strains from the DSMZ strain collection, resulting in the prioritization of 15 potential PSI producer strains. For twelve of them, extract samples showed protein synthesis inhibitory properties in in vitro transcription/translation assays. For four strains, namely Saccharopolyspora flava DSM 44771, Micromonospora aurantiaca DSM 43813, Nocardioides albertanoniae DSM 25218, and Geodermatophilus nigrescens DSM 45408, the protein synthesis inhibitory substance amicoumacin was identified by HPLC-MS analysis, which proved the functionality of the in silico screening approach.
© The Author(s) 2022. Published by Oxford University Press on behalf of NAR Genomics and Bioinformatics.
Figures


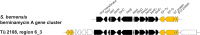


Similar articles
-
Genome sequence of the bialaphos producer Streptomyces sp. DSM 41527 and two putative phosphonate antibiotic producers Streptomyces sp. DSM 41014 and DSM 41981 from the DSMZ strain collection.Access Microbiol. 2024 Apr 19;6(4):000770.v3. doi: 10.1099/acmi.0.000770.v3. eCollection 2024. Access Microbiol. 2024. PMID: 38737806 Free PMC article.
-
Angucycline-like Aromatic Polyketide from a Novel Streptomyces Species Reveals Freshwater Snail Physa acuta as Underexplored Reservoir for Antibiotic-Producing Actinomycetes.Antibiotics (Basel). 2020 Dec 29;10(1):22. doi: 10.3390/antibiotics10010022. Antibiotics (Basel). 2020. PMID: 33383910 Free PMC article.
-
Micromonospora schwarzwaldensis sp. nov., a producer of telomycin, isolated from soil.Int J Syst Evol Microbiol. 2013 Oct;63(Pt 10):3812-3817. doi: 10.1099/ijs.0.051623-0. Epub 2013 May 10. Int J Syst Evol Microbiol. 2013. PMID: 23667141
-
Glycopeptide Antibiotic Resistance Genes: Distribution and Function in the Producer Actinomycetes.Front Microbiol. 2020 Jun 17;11:1173. doi: 10.3389/fmicb.2020.01173. eCollection 2020. Front Microbiol. 2020. PMID: 32655512 Free PMC article. Review.
-
Leveraging a large microbial strain collection for natural product discovery.J Biol Chem. 2019 Nov 8;294(45):16567-16576. doi: 10.1074/jbc.REV119.006514. Epub 2019 Sep 30. J Biol Chem. 2019. PMID: 31570525 Free PMC article. Review.
Cited by
-
Biotransformation-coupled mutasynthesis for the generation of novel pristinamycin derivatives by engineering the phenylglycine residue.RSC Chem Biol. 2023 Oct 14;4(12):1050-1063. doi: 10.1039/d3cb00143a. eCollection 2023 Nov 29. RSC Chem Biol. 2023. PMID: 38033732 Free PMC article.
-
Modern Trends in Natural Antibiotic Discovery.Life (Basel). 2023 Apr 23;13(5):1073. doi: 10.3390/life13051073. Life (Basel). 2023. PMID: 37240718 Free PMC article. Review.
-
Challenging old microbiological treasures for natural compound biosynthesis capacity.Front Bioeng Biotechnol. 2024 Feb 1;12:1255151. doi: 10.3389/fbioe.2024.1255151. eCollection 2024. Front Bioeng Biotechnol. 2024. PMID: 38361790 Free PMC article.
References
-
- WHO Antibiotic Resistance: Multi-country Public Awareness Survey. 2015; World Health Organization.
-
- Lewis K. Platforms for antibiotic discovery. Nat. Rev. Drug Discov. 2013; 12:371–387. - PubMed
-
- Wright G.D. Opportunities for natural products in 21st century antibiotic discovery. Nat. Prod. Rep. 2017; 34:694–701. - PubMed
-
- Wencewicz T.A. New antibiotics from Nature's chemical inventory. Bioorganic Med. Chem. 2016; 24:6227–6252. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

