Comprehensive identification, evolutionary patterns and the divergent response of PRX genes in Phaseolus vulgaris under biotic and abiotic interactions
- PMID: 35855475
- PMCID: PMC9288579
- DOI: 10.1007/s13205-022-03246-8
Comprehensive identification, evolutionary patterns and the divergent response of PRX genes in Phaseolus vulgaris under biotic and abiotic interactions
Abstract
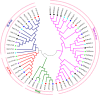
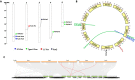
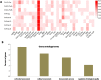
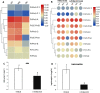
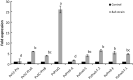
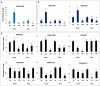
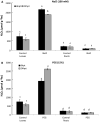
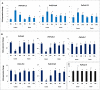
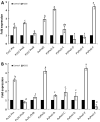
Peroxiredoxins (Prxs) are novel cysteine-based peroxidases which are involved in protecting cells from oxidative damage by catalyzing the reduction of different peroxides. The present study addressed, for the first time, genome-wide identification, evolutionary patterns and expression dynamics of Phaseolus vulgaris Prx gene family (PvPrx). Nine Prx proteins were identified in P. vulgaris based on homology searches. The phylogeny analysis of Prxs from seven plant species revealed that Prx proteins can be clustered into four groups (1C-Prx, 2C-Prxs, PrxQ and type II Prxs). Both tandem and segmental duplication contributed to PvPrx gene family expansion. Intragenic reorganizations including gain/loss of exon/intron and insertions/deletions have also contributed to PvPrx gene diversification. The collinearity analysis revealed the presence of some orthologous Prx gene pairs between A. thaliana and P. vulgaris genomes. The Ka/Ks ratio indicated that two of the three PvPrx duplicated gene pairs have undergone a purifying selection. Redundant stress-related cis-acting elements were also found in the promoters of most PvPrx genes. RT q-PCR analysis revealed an upregulation of key PvPrx members in response to symbiosis and different abiotic factors. The upregulation of targeted PvPrx members, particularly in leaves exposed to salinity or drought, was accompanied by an accumulation of hydrogen peroxide (H2O2). When exogenously applied, H2O2 modulated almost all PvPrx genes, suggesting a potential H2O2-scavenging role for these proteins. Collectively, our analysis provided valuable information for further functional analysis of key PvPrx members to improve common bean stress tolerance and/or its symbiotic performance.
Supplementary information: The online version contains supplementary material available at 10.1007/s13205-022-03246-8.
Keywords: Abiotic stress; Common bean; Peroxiredoxins; Redox regulation; Reverse-transcription q-PCR; Symbiosis.
© King Abdulaziz City for Science and Technology 2022.
Conflict of interest statement
Conflict of interestAll authors declare that they have no conflict of interest.
Figures

References
-
- Ahmad F, Kamal A, Singh A, Ashfaque F, Alamri S, Siddiqui MH. Salicylic acid modulates antioxidant system, defense metabolites, and expression of salt transporter genes in Pisum sativum under salinity stress. J Plant Growth Regul. 2022;41:1905–1918. doi: 10.1007/s00344-020-10271-5. - DOI
-
- Boubakri H, Chihaoui SA, Najjar E, Gargouri M, Barhoumi F, Jebara M. Genome-wide analysis and expression profiling of H-type Trx family in Phaseolus vulgaris revealed distinctive isoforms associated with symbiotic N2-fixing performance and abiotic stress response. J Plant Physiol. 2021;260:153410. doi: 10.1016/j.jplph.2021.153410. - DOI - PubMed
-
- Boubakri H, Saidi MN, Barhoumi F, Jebara M, Brini F (2019) Identification and characterization of thioredoxin h-type gene family in Triticum Turgidium ssp. durum in response to natural and environmental factor-induced oxidative stress. Plant Mol Biol Rep 37(5): 464–476
-
- Broughton WJ, Hernandez G, Blair M, Beebe S, Gepts P, Vanderleyden J. Bean (Phaseolus spp.)—model food legumes. Plant Soil. 2003;25:255–228.
LinkOut - more resources
Full Text Sources

