Infection by Pseudocercospora musae leads to an early reprogramming of the Musa paradisiaca defense transcriptome
- PMID: 35855477
- PMCID: PMC9288577
- DOI: 10.1007/s13205-022-03245-9
Infection by Pseudocercospora musae leads to an early reprogramming of the Musa paradisiaca defense transcriptome
Abstract
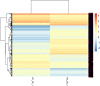
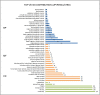
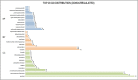
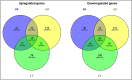
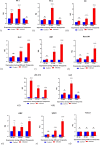
Deep sequencing technologies such as RNA sequencing can help unravel mechanisms governing defense or resistance responses in plant-pathogen interactions. Several studies have been carried out to investigate the transcriptomic changes in Musa germplasm against Yellow Sigatoka disease, but the defense response of Musa paradisiaca has not been investigated so far. We carried out transcriptome sequencing of M. paradisiaca var. Kachkal infected with the pathogen Pseudocercospora musae and found that a vast set of genes were upregulated while many genes were downregulated in the resistant cultivar as a result of infection. After transcriptome assembly and differential gene expression analysis, 429 upregulated and 156 downregulated genes were filtered out (considering fold change ± 2, p < 0.01). Functional annotation of the differentially expressed genes (DEGs) enriched the upregulated genes into 49 gene ontology (GO) classes of biological processes (BP), 20 classes of molecular function (MF) and 9 classes of cellular component (CC). Similarly, the downregulated genes were classified into 35 GO classes of BP, 28 classes of MF and 6 classes of CC. The KEGG enrichment analysis revealed that the upregulated genes were most highly represented in 'metabolic' and 'biosynthesis of secondary metabolites' pathways. Additionally, 'plant hormone signal transduction', 'plant-pathogen interaction' and 'phenylpropanoid biosynthesis' pathways were also significantly enriched indicating their involvement in resistance responses against the pathogen. The RNA-seq analysis also depicts that a range of important defense-related genes are modulated as a result of infection, all of which are responsible for either mediating or activating resistance responses in the host. We studied and validated the expression profiles of ten important defense-related genes potentially involved in conferring resistance to the pathogen through qRT-PCR. Almost all the selected defense-related genes were found to be highly and significantly upregulated within 24 h post inoculation (hpi) and for some genes, the expression remained consistently high till the later time point of 72 hpi. These results, thus, indicate that the infection by P. musae leads to a rapid reprogramming of the defense transcriptome of the resistant banana cultivar. The defense-related genes identified to be modulated in response to infection are important not only for pathogen recognition and perception but also for activation and persistence of defense in the host.
Supplementary information: The online version contains supplementary material available at 10.1007/s13205-022-03245-9.
Keywords: Differentially expressed genes; Host–pathogen interaction; Musa paradisiaca; Sigatoka; Transcriptome.
© King Abdulaziz City for Science and Technology 2022.
Conflict of interest statement
Conflict of interestThe authors declare that they have no conflict of interest.
Figures

References
-
- Aamir M, Singh VK, Dubey MK, Kashyap SP, Zehra A, Upadhyay RS, Singh S. Structural and functional dissection of differentially expressed tomato WRKY transcripts in host defense response against the vascular wilt pathogen (Fusarium oxysporum f. sp. lycopersici) PloS One. 2018 doi: 10.1371/journal.pone.0193922. - DOI - PMC - PubMed
-
- AbuQamar S, Chen X, Dahwan R, Bluhm B, Salmeron J, Lam S, Dietrich RA, Mengiste T. Expression profiling and mutant analysis reveals complex regulatory networks involved in Arabidopsis response to Botrytis infection. Plant J. 2006;48:28–44. - PubMed
-
- Arzanlou M, Abeln E, Kema G, Waalwijk C, Carlier J, de Vries I, Guzmán M, Crous P. Molecular diagnostics for the sigatoka disease complex of banana. Phytopathol. 2007;97:1112–1118. - PubMed
-
- Berrocal-Lobo M, Molina A, Solano R. Constitutive expression of ethylene-response-factor1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J. 2002;29:23–32. - PubMed
LinkOut - more resources
Full Text Sources

