Methods to study folding of alpha-helical membrane proteins in lipids
- PMID: 35855589
- PMCID: PMC9297032
- DOI: 10.1098/rsob.220054
Methods to study folding of alpha-helical membrane proteins in lipids
Abstract
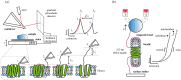
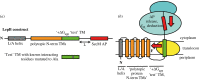
How alpha-helical membrane proteins fold correctly in the highly hydrophobic membrane interior is not well understood. Their folding is known to be highly influenced by the lipids within the surrounding bilayer, but the majority of folding studies have focused on detergent-solubilized protein rather than protein in a lipid environment. There are different ways to study folding in lipid bilayers, and each method has its own advantages and disadvantages. This review will discuss folding methods which can be used to study alpha-helical membrane proteins in bicelles, liposomes, nanodiscs or native membranes. These folding methods include in vitro folding methods in liposomes such as denaturant unfolding studies, and single-molecule force spectroscopy studies in bicelles, liposomes and native membranes. This review will also discuss recent advances in co-translational folding studies, which use cell-free expression with liposomes or nanodiscs or are performed in vivo with native membranes.
Keywords: folding; lipids; membrane; protein.
Conflict of interest statement
The authors declare that there are no competing interests associated with this manuscript.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

