Trained immunity of alveolar macrophages requires metabolic rewiring and type 1 interferon signaling
- PMID: 35856089
- PMCID: PMC9385480
- DOI: 10.1038/s41385-022-00528-5
Trained immunity of alveolar macrophages requires metabolic rewiring and type 1 interferon signaling
Abstract
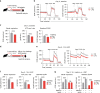
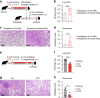
Environmental microbial triggers shape the development and functionality of the immune system. Alveolar macrophages (AMs), tissue-resident macrophages of the lungs, are in constant and direct contact with inhaled particles and microbes. Such exposures likely impact AM reactivity to subsequent challenges by immunological imprinting mechanisms referred to as trained immunity. Here, we investigated whether a ubiquitous microbial compound has the potential to induce AM training in vivo. We discovered that intranasal exposure to ambient amounts of lipopolysaccharide (LPS) induced a pronounced AM memory response, characterized by enhanced reactivity upon pneumococcal challenge. Exploring the mechanistic basis of AM training, we identified a critical role of type 1 interferon signaling and found that inhibition of fatty acid oxidation and glutaminolysis significantly attenuated the training effect. Notably, adoptive transfer of trained AMs resulted in increased bacterial loads and tissue damage upon subsequent pneumococcal infection. In contrast, intranasal pre-exposure to LPS promoted bacterial clearance, highlighting the complexity of stimulus-induced immune responses, which likely involve multiple cell types and may depend on the local immunological and metabolic environment. Collectively, our findings demonstrate the profound impact of ambient microbial exposure on pulmonary immune memory and reveal tissue-specific features of trained immunity.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interest.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

