Evolution of the SARS-CoV-2 omicron variants BA.1 to BA.5: Implications for immune escape and transmission
- PMID: 35856385
- PMCID: PMC9349777
- DOI: 10.1002/rmv.2381
Evolution of the SARS-CoV-2 omicron variants BA.1 to BA.5: Implications for immune escape and transmission
Abstract
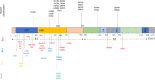
The first dominant SARS-CoV-2 Omicron variant BA.1 harbours 35 mutations in its Spike protein from the original SARS-CoV-2 variant that emerged late 2019. Soon after its discovery, BA.1 rapidly emerged to become the dominant variant worldwide and has since evolved into several variants. Omicron is of major public health concern owing to its high infectivity and antibody evasion. This review article examines the theories that have been proposed on the evolution of Omicron including zoonotic spillage, infection in immunocompromised individuals and cryptic spread in the community without being diagnosed. Added to the complexity of Omicron's evolution are the multiple reports of recombination events occurring between co-circulating variants of Omicron with Delta and other variants such as XE. Current literature suggests that the combination of the novel mutations in Omicron has resulted in the variant having higher infectivity than the original Wuhan-Hu-1 and Delta variant. However, severity is believed to be less owing to the reduced syncytia formation and lower multiplication in the human lung tissue. Perhaps most challenging is that several studies indicate that the efficacy of the available vaccines have been reduced against Omicron variant (8-127 times reduction) as compared to the Wuhan-Hu-1 variant. The administration of booster vaccine, however, compensates with the reduction and improves the efficacy by 12-35 fold. Concerningly though, the broadly neutralising monoclonal antibodies, including those approved by FDA for therapeutic use against previous SARS-CoV-2 variants, are mostly ineffective against Omicron with the exception of Sotrovimab and recent reports suggest that the Omicron BA.2 is also resistant to Sotrovimab. Currently two new Omicron variants BA.4 and BA.5 are emerging and are reported to be more transmissible and resistant to immunity generated by previous variants including Omicron BA.1 and most monoclonal antibodies. As new variants of SARS-CoV-2 will likely continue to emerge it is important that the evolution, and biological consequences of new mutations, in existing variants be well understood.
Keywords: SARS-COV-2; immune evasion; monoclonal antibodies; omicron.
© 2022 The Authors. Reviews in Medical Virology published by John Wiley & Sons Ltd.
Conflict of interest statement
No conflict of interest declared.
Figures

References
-
- World Health Organization. Tracking SARS‐COV‐2 Variants. In.; 2021.
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

