Panoramic comparison between NK cells in healthy and cancerous liver through single-cell RNA sequencing
- PMID: 35856557
- PMCID: PMC9500227
- DOI: 10.20892/j.issn.2095-3941.2022.0050
Panoramic comparison between NK cells in healthy and cancerous liver through single-cell RNA sequencing
Abstract
Objective: NK cells play crucial roles in the immune defense mechanisms against viral infections and transformed cells. However, the developmental progression, transcriptomic landscape, and functional subtypes of liver NK cells are not well defined. Hepatocellular carcinoma (HCC) accounts for approximately 80% of primary liver cancer worldwide, yet the biological characteristics of NK cells in the HCC environment are unclear. Therefore, we aimed to determine these cells' roles in tumorigenesis and prognosis.
Methods: We compared the single-cell RNA sequencing profiles of NK cells purified from blood (n = 1), healthy liver tissues (n = 3), HCC tumor tissues (n = 4), and peritumor liver tissues (n = 1) to identify NK cell subsets. Furthermore, we performed bioinformatics analysis by using The Cancer Genome Atlas (TCGA) data to identify prognostic biomarkers simultaneously overexpressed in the blood and tumor tissues of patients with HCC.
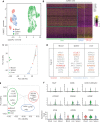
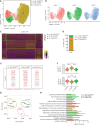
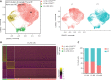
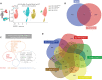
Results: Transcriptomic analysis revealed 5 NK cell subsets (L1-NK-CD56bright, L2-NK-CD56dim, L3-NK-HLA, L4-LrNK-FCGR3A, and L5-LrNK-XCL1) in the healthy liver tissues. However, the transitional L3 subset and the CXCR6+CD16+ L4 subset with strong anti-tumor activity were absent in the HCC and peritumor liver tissues. Furthermore, 4 common prognosis-associated genes (RHOB, TALDO1, HLA-DPA1, and TKT) were significantly overexpressed in the paired tumor tissue and blood.
Conclusions: Our study revealed 5 specific subsets of NK cells in healthy human liver tissues. However, only 3 of the 5 NK cell subsets were present in HCC and peritumor tissues. The cytotoxic NK cell subsets were absent in HCC tissues. Furthermore, we identified 4 potential non-invasive prognostic biomarkers in patients with HCC.
Keywords: Hepatocellular carcinoma; heterogeneity; natural killer cell; prognosis; single-cell RNA sequencing.
Copyright © 2022 Cancer Biology & Medicine.
Conflict of interest statement
No potential conflicts of interest are disclosed.
Figures





References
-
- Vivier E, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, et al. Innate Lymphoid Cells: 10 Years On. Cell. 2018;174:1054–66. - PubMed
-
- Zhang Q, Bi J, Zheng X, Chen Y, Wang H, Wu W, et al. Blockade of the checkpoint receptor TIGIT prevents NK cell exhaustion and elicits potent anti-tumor immunity. Nat Immunol. 2018;19:723–32. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous
