The Gut Bacterial Community Potentiates Clostridioides difficile Infection Severity
- PMID: 35856563
- PMCID: PMC9426473
- DOI: 10.1128/mbio.01183-22
The Gut Bacterial Community Potentiates Clostridioides difficile Infection Severity
Abstract
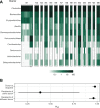
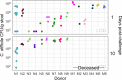
The severity of Clostridioides difficile infections (CDI) has increased over the last few decades. Patient age, white blood cell count, and creatinine levels as well as C. difficile ribotype and toxin genes have been associated with disease severity. However, it is unclear whether specific members of the gut microbiota are associated with variations in disease severity. The gut microbiota is known to interact with C. difficile during infection. Perturbations to the gut microbiota are necessary for C. difficile to colonize the gut. The gut microbiota can inhibit C. difficile colonization through bile acid metabolism, nutrient consumption, and bacteriocin production. Here, we sought to demonstrate that members of the gut bacterial communities can also contribute to disease severity. We derived diverse gut communities by colonizing germfree mice with different human fecal communities. The mice were then infected with a single C. difficile ribotype 027 clinical isolate, which resulted in moribundity and histopathologic differences. The variation in severity was associated with the human fecal community that the mice received. Generally, bacterial populations with pathogenic potential, such as Enterococcus, Helicobacter, and Klebsiella, were associated with more-severe outcomes. Bacterial groups associated with fiber degradation and bile acid metabolism, such as Anaerotignum, Blautia, Lactonifactor, and Monoglobus, were associated with less-severe outcomes. These data indicate that, in addition to the host and C. difficile subtype, populations of gut bacteria can influence CDI disease severity. IMPORTANCE Clostridioides difficile colonization can be asymptomatic or develop into an infection ranging in severity from mild diarrhea to toxic megacolon, sepsis, and death. Models that predict severity and guide treatment decisions are based on clinical factors and C. difficile characteristics. Although the gut microbiome plays a role in protecting against CDI, its effect on CDI disease severity is unclear and has not been incorporated into disease severity models. We demonstrated that variation in the microbiome of mice colonized with human feces yielded a range of disease outcomes. These results revealed groups of bacteria associated with both severe and mild C. difficile infection outcomes. Gut bacterial community data from patients with CDI could improve our ability to identify patients at risk of developing more severe disease and improve interventions that target C. difficile and the gut bacteria to reduce host damage.
Keywords: CDI; Clostridium difficile; human microbiome; humanized mice; microbial ecology.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- McDonald LC, Gerding DN, Johnson S, Bakken JS, Carroll KC, Coffin SE, Dubberke ER, Garey KW, Gould CV, Kelly C, Loo V, Sammons JS, Sandora TJ, Wilcox MH. 2018. Clinical practice guidelines for Clostridium difficile infection in adults and children: 2017 update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA). Clin Infect Dis 66:e1–e48. doi: 10.1093/cid/cix1085. - DOI - PMC - PubMed
-
- Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A, Littmann E, van den Brink MRM, Jenq RR, Taur Y, Sander C, Cross JR, Toussaint NC, Xavier JB, Pamer EG. 2015. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517:205–208. doi: 10.1038/nature13828. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
