Interplay between Sulfur Assimilation and Biodesulfurization Activity in Rhodococcus qingshengii IGTS8: Insights into a Regulatory Role of the Reverse Transsulfuration Pathway
- PMID: 35856606
- PMCID: PMC9426449
- DOI: 10.1128/mbio.00754-22
Interplay between Sulfur Assimilation and Biodesulfurization Activity in Rhodococcus qingshengii IGTS8: Insights into a Regulatory Role of the Reverse Transsulfuration Pathway
Abstract
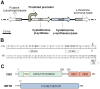
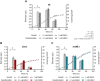
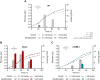
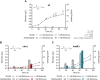
Biodesulfurization is a process that selectively removes sulfur from dibenzothiophene and its derivatives. Several natural biocatalysts harboring the highly conserved desulfurization operon dszABC, which is significantly repressed by methionine, cysteine, and inorganic sulfate, have been isolated. However, the available information on the metabolic regulation of gene expression is still limited. In this study, scarless knockouts of the reverse transsulfuration pathway enzyme genes cbs and metB were constructed in the desulfurizing strain Rhodococcus sp. strain IGTS8. We provide sequence analyses and report the enzymes' involvement in the sulfate- and methionine-dependent repression of biodesulfurization activity. Sulfate addition in the bacterial culture did not repress the desulfurization activity of the Δcbs strain, whereas deletion of metB promoted a significant biodesulfurization activity for sulfate-based growth and an even higher desulfurization activity for methionine-grown cells. In contrast, growth on cysteine completely repressed the desulfurization activity of all strains. Transcript level comparison uncovered a positive effect of cbs and metB gene deletions on dsz gene expression in the presence of sulfate and methionine, but not cysteine, offering insights into a critical role of cystathionine β-synthase (CβS) and MetB in desulfurization activity regulation. IMPORTANCE Precise genome editing of the model biocatalyst Rhodococcus qingshengii IGTS8 was performed for the first time, more than 3 decades after its initial discovery. We thus gained insight into the regulation of dsz gene expression and biocatalyst activity, depending on the presence of two reverse transsulfuration enzymes, CβS and MetB. Moreover, we observed an enhancement of biodesulfurization capability in the presence of otherwise repressive sulfur sources, such as sulfate and l-methionine. The interconnection of cellular sulfur assimilation strategies was revealed and validated.
Keywords: biocatalysis; cystathionine; cystathionine β-synthase; cystathionine γ-lyase; cysteine; genetic engineering; methionine; reverse transsulfuration; sulfur metabolism.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Kilbane JJ. 2017. Biodesulfurization: how to make it work? Arab J Sci Eng 42:1–9. doi:10.1007/s13369-016-2269-1. - DOI
-
- Hirschler A, Carapito C, Maurer L, Zumsteg J, Villette C, Heintz D, Dahl C, Al-Nayal A, Sangal V, Mahmoud H, Van Dorsselaer A, Ismail W. 2021. Biodesulfurization induces reprogramming of sulfur metabolism in Rhodococcus qingshengii IGTS8: proteomics and untargeted metabolomics. Microbiol Spectr 9:e00692-21. doi:10.1128/Spectrum.00692-21. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources

