Impaired Succinate Oxidation Prevents Growth and Influences Drug Susceptibility in Mycobacterium tuberculosis
- PMID: 35856639
- PMCID: PMC9426501
- DOI: 10.1128/mbio.01672-22
Impaired Succinate Oxidation Prevents Growth and Influences Drug Susceptibility in Mycobacterium tuberculosis
Abstract
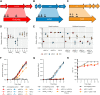
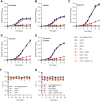
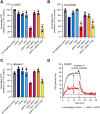
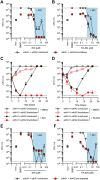
Succinate is a major focal point in mycobacterial metabolism and respiration, serving as both an intermediate of the tricarboxylic acid (TCA) cycle and a direct electron donor for the respiratory chain. Mycobacterium tuberculosis encodes multiple enzymes predicted to be capable of catalyzing the oxidation of succinate to fumarate, including two different succinate dehydrogenases (Sdh1 and Sdh2) and a separate fumarate reductase (Frd) with possible bidirectional behavior. Previous attempts to investigate the essentiality of succinate oxidation in M. tuberculosis have relied on the use of single-gene deletion mutants, raising the possibility that the remaining enzymes could catalyze succinate oxidation in the absence of the other. To address this, we report on the use of mycobacterial CRISPR interference (CRISPRi) to construct single, double, and triple transcriptional knockdowns of sdhA1, sdhA2, and frdA in M. tuberculosis. We show that the simultaneous knockdown of sdhA1 and sdhA2 is required to prevent succinate oxidation and overcome the functional redundancy within these enzymes. Succinate oxidation was demonstrated to be essential for the optimal growth of M. tuberculosis, with the combined knockdown of sdhA1 and sdhA2 significantly impairing the activity of the respiratory chain and preventing growth on a range of carbon sources. Moreover, impaired succinate oxidation was shown to influence the activity of cell wall-targeting antibiotics and bioenergetic inhibitors against M. tuberculosis. Together, these data provide fundamental insights into mycobacterial physiology, energy metabolism, and antimicrobial susceptibility. IMPORTANCE New drugs are urgently required to combat the tuberculosis epidemic that claims 1.5 million lives annually. Inhibitors of mycobacterial energy metabolism have shown significant promise clinically; however, further advancing this nascent target space requires a more fundamental understanding of the respiratory enzymes and pathways used by Mycobacterium tuberculosis. Succinate is a major focal point in mycobacterial metabolism and respiration; yet, the essentiality of succinate oxidation and the consequences of inhibiting this process are poorly defined. In this study, we demonstrate that impaired succinate oxidation prevents the optimal growth of M. tuberculosis on a range of carbon sources and significantly reduces the activity of the electron transport chain. Moreover, we show that impaired succinate oxidation both positively and negatively influences the activity of a variety of antituberculosis drugs. Combined, these findings provide fundamental insights into mycobacterial physiology and drug susceptibility that will be useful in the continued development of bioenergetic inhibitors.
Keywords: CRISPR interference; Mycobacterium tuberculosis; succinate dehydrogenase.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- World Health Organization. 2021. Global tuberculosis report. World Health Organization, Geneva, Switzerland.
-
- World Health Organization. 2010. Guidelines for the treatment of tuberculosis, 4th ed. World Health Organization, Geneva, Switzerland.
-
- Rittershaus ESC, Baek S-H, Krieger IV, Nelson SJ, Cheng Y-S, Nambi S, Baker RE, Leszyk JD, Shaffer SA, Sacchettini JC, Sassetti CM. 2018. A lysine acetyltransferase contributes to the metabolic adaptation to hypoxia in Mycobacterium tuberculosis. Cell Chem Biol 25:1495–1505.e3. doi:10.1016/j.chembiol.2018.09.009. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

