The In Vitro Replication Cycle of Achromobacter xylosoxidans and Identification of Virulence Genes Associated with Cytotoxicity in Macrophages
- PMID: 35856670
- PMCID: PMC9430717
- DOI: 10.1128/spectrum.02083-22
The In Vitro Replication Cycle of Achromobacter xylosoxidans and Identification of Virulence Genes Associated with Cytotoxicity in Macrophages
Abstract
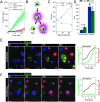
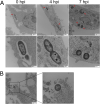
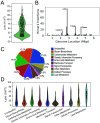
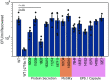
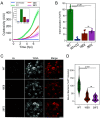
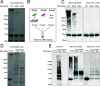
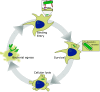
Achromobacter xylosoxidans is an opportunistic pathogen implicated in a wide variety of human infections including the ability to colonize the lungs of cystic fibrosis (CF) patients. The role of A. xylosoxidans in human pathology remains controversial due to the lack of optimized in vitro and in vivo model systems to identify and test bacterial gene products that promote a pathological response. We have previously identified macrophages as a target host cell for A. xylosoxidans-induced cytotoxicity. By optimizing our macrophage infection model, we determined that A. xylosoxidans enters macrophages and can reside within a membrane bound vacuole for extended periods of time. Intracellular replication appears limited with cellular lysis preceding an enhanced, mainly extracellular replication cycle. Using our optimized in vitro model system along with transposon mutagenesis, we identified 163 genes that contribute to macrophage cytotoxicity. From this list, we characterized a giant RTX adhesin encoded downstream of a type one secretion system (T1SS) that mediates bacterial binding and entry into host macrophages, an important first step toward cellular toxicity and inflammation. The RTX adhesin is encoded by other human isolates and is recognized by antibodies present in serum isolated from CF patients colonized by A. xylosoxidans, indicating this virulence factor is produced and deployed in vivo. This study represents the first characterization of A. xylosoxidans replication during infection and identifies a variety of genes that may be linked to virulence and human pathology. IMPORTANCE Patients affected by CF develop chronic bacterial infections characterized by inflammatory exacerbations and tissue damage. Advancements in sequencing technologies have broadened the list of opportunistic pathogens colonizing the CF lung. A. xylosoxidans is increasingly recognized as an opportunistic pathogen in CF, yet our understanding of the bacterium as a contributor to human disease is limited. Genomic studies have identified potential virulence determinants in A. xylosoxidans isolates, but few have been mechanistically studied. Using our optimized in vitro cell model, we identified and characterized a bacterial adhesin that mediates binding and uptake by host macrophages leading to cytotoxicity. A subset of serum samples from CF patients contains antibodies that recognize the RTX adhesion, suggesting, for the first time, that this virulence determinant is produced in vivo. This work furthers our understanding of A. xylosoxidans virulence factors at a mechanistic level.
Keywords: Achromobacter; RTX; cystic fibrosis; cytotoxicity; pathogenesis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Neidhöfer C, Berens C, Parčina M. 2022. An 18-year dataset on the clinical incidence and MICs to antibiotics of Achromobacter spp. (labeled biochemically or by MAL-DI-TOF MS as A. xylosoxidans), largely in patient groups other than those with CF. Antibiotics 11:311. doi: 10.3390/antibiotics11030311. - DOI - PMC - PubMed
-
- Pereira RHV, Leão RS, Carvalho-Assef AP, Albano RM, Rodrigues ERA, Firmida MC, Folescu TW, Plotkowski MC, Bernardo VG, Marques EA. 2017. Patterns of virulence factor expression and antimicrobial resistance in Achromobacter xylosoxidans and Achromobacter ruhlandii isolates from patients with cystic fibrosis. Epidemiol Infect 145:600–606. doi: 10.1017/S0950268816002624. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

