Neuronal activity induces glucosylceramide that is secreted via exosomes for lysosomal degradation in glia
- PMID: 35857503
- PMCID: PMC9278864
- DOI: 10.1126/sciadv.abn3326
Neuronal activity induces glucosylceramide that is secreted via exosomes for lysosomal degradation in glia
Abstract
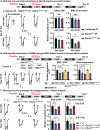
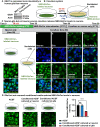
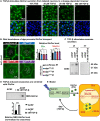
Recessive variants in GBA1 cause Gaucher disease, a prevalent form of lysosome storage disease. GBA1 encodes a lysosomal enzyme that hydrolyzes glucosylceramide (GlcCer) into glucose and ceramide. Its loss causes lysosomal dysfunction and increased levels of GlcCer. We generated a null allele of the Drosophila ortholog Gba1b by inserting the Gal4 using CRISPR-Cas9. Here, we show that Gba1b is expressed in glia but not in neurons. Glial-specific knockdown recapitulates the defects found in Gba1b mutants, and these can be rescued by glial expression of human GBA1. We show that GlcCer is synthesized upon neuronal activity, and it is transported from neurons to glia through exosomes. Furthermore, we found that glial TGF-β/BMP induces the transfer of GlcCer from neurons to glia and that the White protein, an ABCG transporter, promotes GlcCer trafficking to glial lysosomes for degradation.
Figures




Similar articles
-
Lipids regulate the hydrolysis of membrane bound glucosylceramide by lysosomal β-glucocerebrosidase.J Lipid Res. 2017 Mar;58(3):563-577. doi: 10.1194/jlr.M073510. Epub 2017 Jan 26. J Lipid Res. 2017. PMID: 28126847 Free PMC article.
-
Glucocerebrosidase reduces the spread of protein aggregation in a Drosophila melanogaster model of neurodegeneration by regulating proteins trafficked by extracellular vesicles.PLoS Genet. 2021 Feb 4;17(2):e1008859. doi: 10.1371/journal.pgen.1008859. eCollection 2021 Feb. PLoS Genet. 2021. PMID: 33539341 Free PMC article.
-
Glucocerebrosidases catalyze a transgalactosylation reaction that yields a newly-identified brain sterol metabolite, galactosylated cholesterol.J Biol Chem. 2020 Apr 17;295(16):5257-5277. doi: 10.1074/jbc.RA119.012502. Epub 2020 Mar 6. J Biol Chem. 2020. PMID: 32144204 Free PMC article.
-
Lysosomal functions and dysfunctions: Molecular and cellular mechanisms underlying Gaucher disease and its association with Parkinson disease.Adv Drug Deliv Rev. 2022 Aug;187:114402. doi: 10.1016/j.addr.2022.114402. Epub 2022 Jun 25. Adv Drug Deliv Rev. 2022. PMID: 35764179 Review.
-
The metabolism of glucocerebrosides - From 1965 to the present.Mol Genet Metab. 2017 Jan-Feb;120(1-2):22-26. doi: 10.1016/j.ymgme.2016.11.390. Epub 2016 Dec 2. Mol Genet Metab. 2017. PMID: 27955980 Review.
Cited by
-
Sphingolipids in neurodegenerative diseases.Front Neurosci. 2023 Feb 16;17:1137893. doi: 10.3389/fnins.2023.1137893. eCollection 2023. Front Neurosci. 2023. PMID: 36875645 Free PMC article. Review.
-
Ups and downs of lysosomal pH: conflicting roles of LAMP proteins?Autophagy. 2024 Feb;20(2):437-440. doi: 10.1080/15548627.2023.2274253. Epub 2024 Jan 25. Autophagy. 2024. PMID: 37960894 Free PMC article.
-
Exploring therapeutic strategies for infantile neuronal axonal dystrophy (INAD/PARK14).Elife. 2023 Jan 16;12:e82555. doi: 10.7554/eLife.82555. Elife. 2023. PMID: 36645408 Free PMC article.
-
Deletion of Gba in neurons, but not microglia, causes neurodegeneration in a Gaucher mouse model.JCI Insight. 2024 Nov 8;9(21):e179126. doi: 10.1172/jci.insight.179126. JCI Insight. 2024. PMID: 39312723 Free PMC article.
-
Glia-derived secretory fatty acid binding protein Obp44a regulates lipid storage and efflux in the developing Drosophila brain.bioRxiv [Preprint]. 2024 Apr 11:2024.04.10.588417. doi: 10.1101/2024.04.10.588417. bioRxiv. 2024. PMID: 38645138 Free PMC article. Preprint.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

