Pluripotency factors regulate the onset of Hox cluster activation in the early embryo
- PMID: 35857513
- PMCID: PMC9286507
- DOI: 10.1126/sciadv.abo3583
Pluripotency factors regulate the onset of Hox cluster activation in the early embryo
Abstract
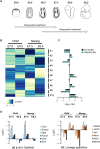
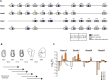
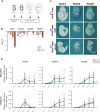
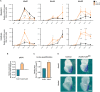
Pluripotent cells are a transient population of the mammalian embryo dependent on transcription factors, such as OCT4 and NANOG, which maintain pluripotency while suppressing lineage specification. However, these factors are also expressed during early phases of differentiation, and their role in the transition from pluripotency to lineage specification is largely unknown. We found that pluripotency factors play a dual role in regulating key lineage specifiers, initially repressing their expression and later being required for their proper activation. We show that Oct4 is necessary for activation of HoxB genes during differentiation of embryonic stem cells and in the embryo. In addition, we show that the HoxB cluster is coordinately regulated by OCT4 binding sites located at the 3' end of the cluster. Our results show that core pluripotency factors are not limited to maintaining the precommitted epiblast but are also necessary for the proper deployment of subsequent developmental programs.
Figures


References
-
- Downs K. M., Systematic localization of Oct-3/4 to the gastrulating mouse conceptus suggests manifold roles in mammalian development. Dev. Dyn. 237, 464–475 (2008). - PubMed
-
- Hart A. H., Hartley L., Ibrahim M., Robb L., Identification, cloning and expression analysis of the pluripotency promoting Nanog genes in mouse and human. Dev. Dyn. 230, 187–198 (2004). - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

