Omicron spike function and neutralizing activity elicited by a comprehensive panel of vaccines
- PMID: 35857529
- PMCID: PMC9348749
- DOI: 10.1126/science.abq0203
Omicron spike function and neutralizing activity elicited by a comprehensive panel of vaccines
Abstract
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Omicron variant of concern comprises several sublineages, with BA.2 and BA.2.12.1 having replaced the previously dominant BA.1 and with BA.4 and BA.5 increasing in prevalence worldwide. We show that the large number of Omicron sublineage spike mutations leads to enhanced angiotensin-converting enzyme 2 (ACE2) binding, reduced fusogenicity, and severe dampening of plasma neutralizing activity elicited by infection or seven clinical vaccines relative to the ancestral virus. Administration of a homologous or heterologous booster based on the Wuhan-Hu-1 spike sequence markedly increased neutralizing antibody titers and breadth against BA.1, BA.2, BA.2.12.1, BA.4, and BA.5 across all vaccines evaluated. Our data suggest that although Omicron sublineages evade polyclonal neutralizing antibody responses elicited by primary vaccine series, vaccine boosters may provide sufficient protection against Omicron-induced severe disease.
Figures
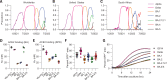
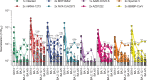
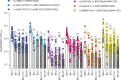
Update of
-
Omicron BA.1 and BA.2 neutralizing activity elicited by a comprehensive panel of human vaccines.bioRxiv [Preprint]. 2022 Mar 16:2022.03.15.484542. doi: 10.1101/2022.03.15.484542. bioRxiv. 2022. Update in: Science. 2022 Aug 19;377(6608):890-894. doi: 10.1126/science.abq0203. PMID: 35313570 Free PMC article. Updated. Preprint.
Comment in
-
Omicron spike protein: a clue for viral entry and immune evasion.Signal Transduct Target Ther. 2022 Sep 28;7(1):339. doi: 10.1038/s41392-022-01193-7. Signal Transduct Target Ther. 2022. PMID: 36171200 Free PMC article. No abstract available.
References
-
- McCallum M., Bassi J., De Marco A., Chen A., Walls A. C., Di Iulio J., Tortorici M. A., Navarro M.-J., Silacci-Fregni C., Saliba C., Sprouse K. R., Agostini M., Pinto D., Culap K., Bianchi S., Jaconi S., Cameroni E., Bowen J. E., Tilles S. W., Pizzuto M. S., Guastalla S. B., Bona G., Pellanda A. F., Garzoni C., Van Voorhis W. C., Rosen L. E., Snell G., Telenti A., Virgin H. W., Piccoli L., Corti D., Veesler D., SARS-CoV-2 immune evasion by the B.1.427/B.1.429 variant of concern. Science 373, 648–654 (2021). 10.1126/science.abi7994 - DOI - PMC - PubMed
-
- Hodcroft E. B., Zuber M., Nadeau S., Vaughan T. G., Crawford K. H. D., Althaus C. L., Reichmuth M. L., Bowen J. E., Walls A. C., Corti D., Bloom J. D., Veesler D., Mateo D., Hernando A., Comas I., González-Candelas F., SeqCOVID-SPAIN consortium, Stadler T., Neher R. A., Spread of a SARS-CoV-2 variant through Europe in the summer of 2020. Nature 595, 707–712 (2021). 10.1038/s41586-021-03677-y - DOI - PubMed
-
- McCallum M., Walls A. C., Sprouse K. R., Bowen J. E., Rosen L. E., Dang H. V., De Marco A., Franko N., Tilles S. W., Logue J., Miranda M. C., Ahlrichs M., Carter L., Snell G., Pizzuto M. S., Chu H. Y., Van Voorhis W. C., Corti D., Veesler D., Molecular basis of immune evasion by the Delta and Kappa SARS-CoV-2 variants. Science 374, 1621–1626 (2021). 10.1126/science.abl8506 - DOI - PMC - PubMed
-
- McCallum M., Czudnochowski N., Rosen L. E., Zepeda S. K., Bowen J. E., Walls A. C., Hauser K., Joshi A., Stewart C., Dillen J. R., Powell A. E., Croll T. I., Nix J., Virgin H. W., Corti D., Snell G., Veesler D., Structural basis of SARS-CoV-2 Omicron immune evasion and receptor engagement. Science 375, 864–868 (2022). 10.1126/science.abn8652 - DOI - PMC - PubMed
-
- Walls A. C., Sprouse K. R., Bowen J. E., Joshi A., Franko N., Navarro M. J., Stewart C., Cameroni E., McCallum M., Goecker E. A., Degli-Angeli E. J., Logue J., Greninger A., Corti D., Chu H. Y., Veesler D., SARS-CoV-2 breakthrough infections elicit potent, broad, and durable neutralizing antibody responses. Cell 185, 872–880.e3 (2022). 10.1016/j.cell.2022.01.011 - DOI - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

