Immunoglobulin A antibody composition is sculpted to bind the self gut microbiome
- PMID: 35857580
- PMCID: PMC9421563
- DOI: 10.1126/sciimmunol.abg3208
Immunoglobulin A antibody composition is sculpted to bind the self gut microbiome
Abstract
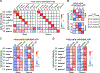
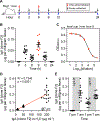
Despite being the most abundantly secreted immunoglobulin isotype, the pattern of reactivity of immunoglobulin A (IgA) antibodies toward each individual's own gut commensal bacteria still remains elusive. By colonizing germ-free mice with defined commensal bacteria, we found that the binding specificity of bulk fecal and serum IgA toward resident gut bacteria resolves well at the species level and has modest strain-level specificity. IgA hybridomas generated from lamina propria B cells of gnotobiotic mice showed that most IgA clones recognized a single bacterial species, whereas a small portion displayed cross-reactivity. Orally administered hybridoma-produced IgAs still retained bacterial antigen binding capability, implying the potential for a new class of therapeutic antibodies. Species-specific IgAs had a range of strain specificities. Given the distinctive bacterial species and strain composition found in each individual's gut, our findings suggest the IgA antibody repertoire is shaped uniquely to bind "self" gut bacteria.
Conflict of interest statement
Figures


References
-
- Sutherland DB, Suzuki K, Fagarasan S, Fostering of advanced mutualism with gut microbiota by Immunoglobulin A. Immunological Reviews 270, 20–31 (2016). - PubMed
-
- Macpherson AJ, Geuking MB, McCoy KD, Homeland Security: IgA immunity at the frontiers of the body. Trends Immunol 33, 160–167 (2012). - PubMed
-
- Yang Y, Palm NW, Immunoglobulin A and the microbiome. Curr Opin Microbiol 56, 89–96 (2020). - PubMed
-
- Hand TW, Reboldi A, Production and Function of Immunoglobulin A. Annu Rev Immunol 39, 695–718 (2021). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

