Mosaic RBD nanoparticles protect against challenge by diverse sarbecoviruses in animal models
- PMID: 35857620
- PMCID: PMC9273039
- DOI: 10.1126/science.abq0839
Mosaic RBD nanoparticles protect against challenge by diverse sarbecoviruses in animal models
Abstract
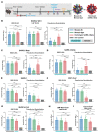
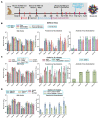
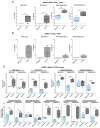
To combat future severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants and spillovers of SARS-like betacoronaviruses (sarbecoviruses) threatening global health, we designed mosaic nanoparticles that present randomly arranged sarbecovirus spike receptor-binding domains (RBDs) to elicit antibodies against epitopes that are conserved and relatively occluded rather than variable, immunodominant, and exposed. We compared immune responses elicited by mosaic-8 (SARS-CoV-2 and seven animal sarbecoviruses) and homotypic (only SARS-CoV-2) RBD nanoparticles in mice and macaques and observed stronger responses elicited by mosaic-8 to mismatched (not on nanoparticles) strains, including SARS-CoV and animal sarbecoviruses. Mosaic-8 immunization showed equivalent neutralization of SARS-CoV-2 variants, including Omicrons, and protected from SARS-CoV-2 and SARS-CoV challenges, whereas homotypic SARS-CoV-2 immunization protected only from SARS-CoV-2 challenge. Epitope mapping demonstrated increased targeting of conserved epitopes after mosaic-8 immunization. Together, these results suggest that mosaic-8 RBD nanoparticles could protect against SARS-CoV-2 variants and future sarbecovirus spillovers.
Figures



Update of
-
Mosaic RBD nanoparticles protect against multiple sarbecovirus challenges in animal models.bioRxiv [Preprint]. 2022 Mar 28:2022.03.25.485875. doi: 10.1101/2022.03.25.485875. bioRxiv. 2022. Update in: Science. 2022 Aug 5;377(6606):eabq0839. doi: 10.1126/science.abq0839. PMID: 35378752 Free PMC article. Updated. Preprint.
References
-
- Planas D., Bruel T., Grzelak L., Guivel-Benhassine F., Staropoli I., Porrot F., Planchais C., Buchrieser J., Rajah M. M., Bishop E., Albert M., Donati F., Prot M., Behillil S., Enouf V., Maquart M., Smati-Lafarge M., Varon E., Schortgen F., Yahyaoui L., Gonzalez M., De Sèze J., Péré H., Veyer D., Sève A., Simon-Lorière E., Fafi-Kremer S., Stefic K., Mouquet H., Hocqueloux L., van der Werf S., Prazuck T., Schwartz O., Sensitivity of infectious SARS-CoV-2 B.1.1.7 and B.1.351 variants to neutralizing antibodies. Nat. Med. 27, 917–924 (2021). 10.1038/s41591-021-01318-5 - DOI - PubMed
-
- Washington N. L., Gangavarapu K., Zeller M., Bolze A., Cirulli E. T., Schiabor Barrett K. M., Larsen B. B., Anderson C., White S., Cassens T., Jacobs S., Levan G., Nguyen J., Ramirez J. M. 3rd, Rivera-Garcia C., Sandoval E., Wang X., Wong D., Spencer E., Robles-Sikisaka R., Kurzban E., Hughes L. D., Deng X., Wang C., Servellita V., Valentine H., De Hoff P., Seaver P., Sathe S., Gietzen K., Sickler B., Antico J., Hoon K., Liu J., Harding A., Bakhtar O., Basler T., Austin B., MacCannell D., Isaksson M., Febbo P. G., Becker D., Laurent M., McDonald E., Yeo G. W., Knight R., Laurent L. C., de Feo E., Worobey M., Chiu C. Y., Suchard M. A., Lu J. T., Lee W., Andersen K. G., Emergence and rapid transmission of SARS-CoV-2 B.1.1.7 in the United States. Cell 184, 2587–2594.e7 (2021). 10.1016/j.cell.2021.03.052 - DOI - PMC - PubMed
-
- Liu L., Iketani S., Guo Y., Chan J. F.-W., Wang M., Liu L., Luo Y., Chu H., Huang Y., Nair M. S., Yu J., Chik K. K.-H., Yuen T. T.-T., Yoon C., To K. K.-W., Chen H., Yin M. T., Sobieszczyk M. E., Huang Y., Wang H. H., Sheng Z., Yuen K.-Y., Ho D. D., Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature 602, 676–681 (2022). 10.1038/s41586-021-04388-0 - DOI - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

