EBNA2-EBF1 complexes promote MYC expression and metabolic processes driving S-phase progression of Epstein-Barr virus-infected B cells
- PMID: 35857872
- PMCID: PMC9335265
- DOI: 10.1073/pnas.2200512119
EBNA2-EBF1 complexes promote MYC expression and metabolic processes driving S-phase progression of Epstein-Barr virus-infected B cells
Abstract
Epstein-Barr virus (EBV) is a human tumor virus which preferentially infects resting human B cells. Upon infection in vitro, EBV activates and immortalizes these cells. The viral latent protein EBV nuclear antigen 2 (EBNA2) is essential for B cell activation and immortalization; it targets and binds the cellular and ubiquitously expressed DNA-binding protein CBF1, thereby transactivating a plethora of viral and cellular genes. In addition, EBNA2 uses its N-terminal dimerization (END) domain to bind early B cell factor 1 (EBF1), a pioneer transcription factor specifying the B cell lineage. We found that EBNA2 exploits EBF1 to support key metabolic processes and to foster cell cycle progression of infected B cells in their first cell cycles upon activation. The α1-helix within the END domain was found to promote EBF1 binding. EBV mutants lacking the α1-helix in EBNA2 can infect and activate B cells efficiently, but activated cells fail to complete the early S phase of their initial cell cycle. Expression of MYC, target genes of MYC and E2F, as well as multiple metabolic processes linked to cell cycle progression are impaired in EBVΔα1-infected B cells. Our findings indicate that EBF1 controls B cell activation via EBNA2 and, thus, has a critical role in regulating the cell cycle of EBV-infected B cells. This is a function of EBF1 going beyond its well-known contribution to B cell lineage specification.
Keywords: B cell activation; Epstein-Barr virus; MYC expression; RNA sequencing; transcription factor.
Conflict of interest statement
The authors declare no competing interest.
Figures

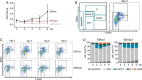
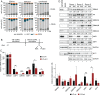
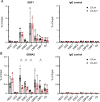

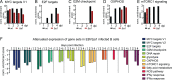
Similar articles
-
Epstein-Barr virus exploits intrinsic B-lymphocyte transcription programs to achieve immortal cell growth.Proc Natl Acad Sci U S A. 2011 Sep 6;108(36):14902-7. doi: 10.1073/pnas.1108892108. Epub 2011 Jul 11. Proc Natl Acad Sci U S A. 2011. PMID: 21746931 Free PMC article.
-
EBF1 binds to EBNA2 and promotes the assembly of EBNA2 chromatin complexes in B cells.PLoS Pathog. 2017 Oct 2;13(10):e1006664. doi: 10.1371/journal.ppat.1006664. eCollection 2017 Oct. PLoS Pathog. 2017. PMID: 28968461 Free PMC article.
-
Enhancer Control of MicroRNA miR-155 Expression in Epstein-Barr Virus-Infected B Cells.J Virol. 2018 Sep 12;92(19):e00716-18. doi: 10.1128/JVI.00716-18. Print 2018 Oct 1. J Virol. 2018. PMID: 30021904 Free PMC article.
-
EBNA2 and Its Coactivator EBNA-LP.Curr Top Microbiol Immunol. 2015;391:35-59. doi: 10.1007/978-3-319-22834-1_2. Curr Top Microbiol Immunol. 2015. PMID: 26428371 Review.
-
EBNA2 and Notch signalling in Epstein-Barr virus mediated immortalization of B lymphocytes.Semin Cancer Biol. 2001 Dec;11(6):423-34. doi: 10.1006/scbi.2001.0409. Semin Cancer Biol. 2001. PMID: 11669604 Review.
Cited by
-
EZH2-Myc Hallmark in Oncovirus/Cytomegalovirus Infections and Cytomegalovirus' Resemblance to Oncoviruses.Cells. 2024 Mar 19;13(6):541. doi: 10.3390/cells13060541. Cells. 2024. PMID: 38534385 Free PMC article. Review.
-
Epstein-Barr Virus B Cell Growth Transformation: The Nuclear Events.Viruses. 2023 Mar 24;15(4):832. doi: 10.3390/v15040832. Viruses. 2023. PMID: 37112815 Free PMC article. Review.
-
EBNA leader protein orchestrates chromatin architecture remodeling during Epstein-Barr virus-induced B cell transformation.Nucleic Acids Res. 2025 Jun 20;53(12):gkaf629. doi: 10.1093/nar/gkaf629. Nucleic Acids Res. 2025. PMID: 40598900 Free PMC article.
-
PSMB1 Inhibits the Replication of Porcine Reproductive and Respiratory Syndrome Virus by Recruiting NBR1 To Degrade Nonstructural Protein 12 by Autophagy.J Virol. 2023 Jan 31;97(1):e0166022. doi: 10.1128/jvi.01660-22. Epub 2023 Jan 5. J Virol. 2023. PMID: 36602366 Free PMC article.
-
Identification and catalog of viral transcriptional regulators in human diseases.iScience. 2025 Feb 21;28(3):112081. doi: 10.1016/j.isci.2025.112081. eCollection 2025 Mar 21. iScience. 2025. PMID: 40124487 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

