An unexplored angle: T cell antigen discoveries reveal a marginal contribution of proteasome splicing to the immunogenic MHC class I antigen pool
- PMID: 35858315
- PMCID: PMC9303865
- DOI: 10.1073/pnas.2119736119
An unexplored angle: T cell antigen discoveries reveal a marginal contribution of proteasome splicing to the immunogenic MHC class I antigen pool
Abstract
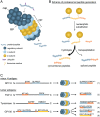
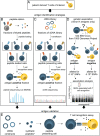
In the current era of T cell-based immunotherapies, it is crucial to understand which types of MHC-presented T cell antigens are produced by tumor cells. In addition to linear peptide antigens, chimeric peptides are generated through proteasome-catalyzed peptide splicing (PCPS). Whether such spliced peptides are abundantly presented by MHC is highly disputed because of disagreement in computational analyses of mass spectrometry data of MHC-eluted peptides. Moreover, such mass spectrometric analyses cannot elucidate how much spliced peptides contribute to the pool of immunogenic antigens. In this Perspective, we explain the significance of knowing the contribution of spliced peptides for accurate analyses of peptidomes on one hand, and to serve as a potential source of targetable tumor antigens on the other hand. Toward a strategy for mass spectrometry independent estimation of the contribution of PCPS to the immunopeptidome, we first reviewed methodologies to identify MHC-presented spliced peptide antigens expressed by tumors. Data from these identifications allowed us to compile three independent datasets containing 103, 74, and 83 confirmed T cell antigens from cancer patients. Only 3.9%, 1.4%, and between 0% and 7.2% of these truly immunogenic antigens are produced by PCPS, therefore providing a marginal contribution to the pool of immunogenic tumor antigens. We conclude that spliced peptides will not serve as a comprehensive source to expand the number of targetable antigens for immunotherapies.
Keywords: PCPS; spliced antigens; tumor antigens.
Conflict of interest statement
Competing interest statement: R.M.S. now works as principal scientist at Neogene Therapeutics BV, which may be perceived as a competing interest.
Figures

Similar articles
-
CD8(+) T cells of Listeria monocytogenes-infected mice recognize both linear and spliced proteasome products.Eur J Immunol. 2016 May;46(5):1109-18. doi: 10.1002/eji.201545989. Epub 2016 Mar 16. Eur J Immunol. 2016. PMID: 26909514
-
Proteasome-Generated cis-Spliced Peptides and Their Potential Role in CD8+ T Cell Tolerance.Front Immunol. 2021 Feb 24;12:614276. doi: 10.3389/fimmu.2021.614276. eCollection 2021. Front Immunol. 2021. PMID: 33717099 Free PMC article.
-
A large fraction of HLA class I ligands are proteasome-generated spliced peptides.Science. 2016 Oct 21;354(6310):354-358. doi: 10.1126/science.aaf4384. Epub 2016 Oct 20. Science. 2016. PMID: 27846572
-
Post-Translational Peptide Splicing and T Cell Responses.Trends Immunol. 2017 Dec;38(12):904-915. doi: 10.1016/j.it.2017.07.011. Epub 2017 Aug 19. Trends Immunol. 2017. PMID: 28830734 Review.
-
Peptide splicing by the proteasome.J Biol Chem. 2017 Dec 22;292(51):21170-21179. doi: 10.1074/jbc.R117.807560. Epub 2017 Nov 6. J Biol Chem. 2017. PMID: 29109146 Free PMC article. Review.
Cited by
-
Identification of T cell antigens in the 21st century, as difficult as ever.Semin Immunol. 2022 Mar;60:101659. doi: 10.1016/j.smim.2022.101659. Epub 2022 Sep 29. Semin Immunol. 2022. PMID: 36183497 Free PMC article. Review.
-
Exploring the Immunogenicity of Noncanonical HLA-I Tumor Ligands Identified through Proteogenomics.Clin Cancer Res. 2023 Jun 13;29(12):2250-2265. doi: 10.1158/1078-0432.CCR-22-3298. Clin Cancer Res. 2023. PMID: 36749875 Free PMC article.
-
Computation strategies and clinical applications in neoantigen discovery towards precision cancer immunotherapy.Biomark Res. 2025 Jul 9;13(1):96. doi: 10.1186/s40364-025-00808-9. Biomark Res. 2025. PMID: 40629481 Free PMC article. Review.
-
Immunogenic self-peptides - the great unknowns in autoimmunity: Identifying T-cell epitopes driving the autoimmune response in autoimmune diseases.Front Immunol. 2023 Jan 9;13:1097871. doi: 10.3389/fimmu.2022.1097871. eCollection 2022. Front Immunol. 2023. PMID: 36700227 Free PMC article. Review.
References
-
- Warren E. H., et al. , An antigen produced by splicing of noncontiguous peptides in the reverse order. Science 313, 1444–1447 (2006). - PubMed
-
- Hanada K., Yewdell J. W., Yang J. C., Immune recognition of a human renal cancer antigen through post-translational protein splicing. Nature 427, 252–256 (2004). - PubMed
-
- Vigneron N., Ferrari V., Stroobant V., Habib J. A., Van Den Eynde B. J., An antigenic peptide produced by peptide splicing in the proteasome. Science 304, 587–590 (2004). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

