Mass spectrometry imaging to explore molecular heterogeneity in cell culture
- PMID: 35858333
- PMCID: PMC9303856
- DOI: 10.1073/pnas.2114365119
Mass spectrometry imaging to explore molecular heterogeneity in cell culture
Abstract
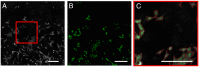
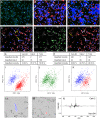
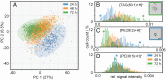
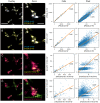
Molecular analysis on the single-cell level represents a rapidly growing field in the life sciences. While bulk analysis from a pool of cells provides a general molecular profile, it is blind to heterogeneities between individual cells. This heterogeneity, however, is an inherent property of every cell population. Its analysis is fundamental to understanding the development, function, and role of specific cells of the same genotype that display different phenotypical properties. Single-cell mass spectrometry (MS) aims to provide broad molecular information for a significantly large number of cells to help decipher cellular heterogeneity using statistical analysis. Here, we present a sensitive approach to single-cell MS based on high-resolution MALDI-2-MS imaging in combination with MALDI-compatible staining and use of optical microscopy. Our approach allowed analyzing large amounts of unperturbed cells directly from the growth chamber. Confident coregistration of both modalities enabled a reliable compilation of single-cell mass spectra and a straightforward inclusion of optical as well as mass spectrometric features in the interpretation of data. The resulting multimodal datasets permit the use of various statistical methods like machine learning-driven classification and multivariate analysis based on molecular profile and establish a direct connection of MS data with microscopy information of individual cells. Displaying data in the form of histograms for individual signal intensities helps to investigate heterogeneous expression of specific lipids within the cell culture and to identify subpopulations intuitively. Ultimately, t-MALDI-2-MSI measurements at 2-µm pixel sizes deliver a glimpse of intracellular lipid distributions and reveal molecular profiles for subcellular domains.
Keywords: cellular heterogeneity; lipidomics; single-cell mass spectrometry; t-MALDI-2-MSI.
Conflict of interest statement
The authors declare no competing interest.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

