A general theory for temperature dependence in biology
- PMID: 35858416
- PMCID: PMC9335213
- DOI: 10.1073/pnas.2119872119
A general theory for temperature dependence in biology
Abstract
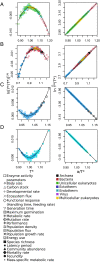
At present, there is no simple, first principles-based, and general model for quantitatively describing the full range of observed biological temperature responses. Here we derive a general theory for temperature dependence in biology based on Eyring-Evans-Polanyi's theory for chemical reaction rates. Assuming only that the conformational entropy of molecules changes with temperature, we derive a theory for the temperature dependence of enzyme reaction rates which takes the form of an exponential function modified by a power law and that describes the characteristic asymmetric curved temperature response. Based on a few additional principles, our model can be used to predict the temperature response above the enzyme level, thus spanning quantum to classical scales. Our theory provides an analytical description for the shape of temperature response curves and demonstrates its generality by showing the convergence of all temperature dependence responses onto universal relationships-a universal data collapse-under appropriate normalization and by identifying a general optimal temperature, around 25 ∘C, characterizing all temperature response curves. The model provides a good fit to empirical data for a wide variety of biological rates, times, and steady-state quantities, from molecular to ecological scales and across multiple taxonomic groups (from viruses to mammals). This theory provides a simple framework to understand and predict the impact of temperature on biological quantities based on the first principles of thermodynamics, bridging quantum to classical scales.
Keywords: metabolic theory; scaling; temperature kinetics.
Conflict of interest statement
The authors declare no competing interest.
Figures

References
-
- Hochachka P. W., Somero G. N., Biochemical Adaptation: Mechanism and Process in Physiological Evolution (Oxford University Press, 2002).
-
- Brown J. H., Gillooly J. F., Allen A. P., Savage V. M., West G. B., Toward a metabolic theory of ecology. Ecology 85, 1771–1789 (2004).
-
- Schulte P. M., Healy T. M., Fangue N. A., Thermal performance curves, phenotypic plasticity, and the time scales of temperature exposure. Integr. Comp. Biol. 51, 691–702 (2011). - PubMed
-
- Grimaud G. M., Mairet F., Sciandra A., Bernard O., Modeling the temperature effect on the specific growth rate of phytoplankton: A review. Rev. Environ. Sci. Biotechnol. 16, 625–645 (2017).
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

