Seed DNA damage responses promote germination and growth in Arabidopsis thaliana
- PMID: 35858436
- PMCID: PMC9335332
- DOI: 10.1073/pnas.2202172119
Seed DNA damage responses promote germination and growth in Arabidopsis thaliana
Abstract
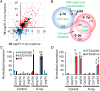
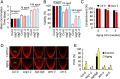
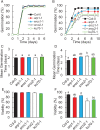
The desiccated, quiescent state of seeds confers extended survival of the embryonic plant. However, accumulation of striking levels of genome damage in quiescence impairs germination and threatens plant survival. The mechanisms by which seeds mitigate this damage remain unclear. Here, we reveal that imbibed Arabidopsis seeds display high resistance to DNA damage, which is lost as seeds advance to germination, coincident with increasing cell cycle activity. In contrast to seedlings, we show that seeds minimize the impact of DNA damage by reducing meristem disruption and delaying SOG1-dependent programmed cell death. This promotes root growth early postgermination. In response to naturally accumulated DNA damage in aging seeds, SOG1 activates cell death postgermination. SOG1 activities are also important for promoting successful seedling establishment. These distinct cellular responses of seeds and seedlings are reflected by different DNA damage transcriptional profiles. Comparative analysis of DNA repair mutants identifies roles of the major genome maintenance pathways in germination but that the repair of cytotoxic chromosomal breaks is the most important for seed longevity. Collectively, these results indicate that high levels of DNA damage incurred in seeds are countered by low cell cycle activity, cell cycle checkpoints, and DNA repair, promoting successful seedling establishment. Our findings reveal insight into both the physiological significance of plant DNA damage responses and the mechanisms which maintain seed longevity, important for survival of plant populations in the natural environment and sustainable crop production under changing climates.
Keywords: DNA repair; genome stability; germination; seed; seed quality.
Conflict of interest statement
The authors declare no competing interest.
Figures

References
-
- Pedroza-Garcia J. A., Xiang Y., De Veylder L., Cell cycle checkpoint control in response to DNA damage by environmental stresses. Plant J. 109, 490–507 (2022). - PubMed
-
- Culligan K. M., Robertson C. E., Foreman J., Doerner P., Britt A. B., ATR and ATM play both distinct and additive roles in response to ionizing radiation. Plant J. 48, 947–961 (2006). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

