Light regulates xylem cell differentiation via PIF in Arabidopsis
- PMID: 35858547
- PMCID: PMC9638722
- DOI: 10.1016/j.celrep.2022.111075
Light regulates xylem cell differentiation via PIF in Arabidopsis
Abstract
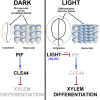
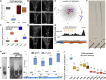
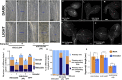
The balance between cell proliferation and differentiation in the cambium defines the formation of plant vascular tissues. As cambium cells proliferate, subsets of daughter cells differentiate into xylem or phloem. TDIF-PXY/TDR signaling is central to this process. TDIF, encoded by CLE41 and CLE44, activates PXY/TDR receptors to maintain proliferative cambium. Light and water are necessary for photosynthesis; thus, vascular differentiation must occur upon light perception to facilitate the transport of water and minerals to the photosynthetic tissues. However, the molecular mechanism controlling vascular differentiation in response to light remains elusive. In this study we show that the accumulation of PIF transcription factors in the dark promotes TDIF signaling and inhibits vascular cell differentiation. On the contrary, PIF inactivation by light leads to a decay in TDIF activity, which induces vascular cell differentiation. Our study connects light to vascular differentiation and highlights the importance of this crosstalk to fine-tune water transport.
Keywords: Arabidopsis; CP; PIF; Plants; cell differentiation; light signaling; photomorphogenesis; plant development; signaling transduction; transcriptional regulation; vascular development; xylem.
Copyright © 2022 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

