Radiogenomic analysis reveals tumor heterogeneity of triple-negative breast cancer
- PMID: 35858585
- PMCID: PMC9381418
- DOI: 10.1016/j.xcrm.2022.100694
Radiogenomic analysis reveals tumor heterogeneity of triple-negative breast cancer
Abstract
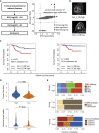
Triple-negative breast cancer (TNBC) is a subset of breast cancer with an adverse prognosis and significant tumor heterogeneity. Here, we extract quantitative radiomic features from contrast-enhanced magnetic resonance images to construct a breast cancer radiomic dataset (n = 860) and a TNBC radiogenomic dataset (n = 202). We develop and validate radiomic signatures that can fairly differentiate TNBC from other breast cancer subtypes and distinguish molecular subtypes within TNBC. A radiomic feature that captures peritumoral heterogeneity is determined to be a prognostic factor for recurrence-free survival (p = 0.01) and overall survival (p = 0.004) in TNBC. Combined with the established matching TNBC transcriptomic and metabolomic data, we demonstrate that peritumoral heterogeneity is associated with immune suppression and upregulated fatty acid synthesis in tumor samples. Collectively, this multi-omic dataset serves as a useful public resource to promote precise subtyping of TNBC and helps to understand the biological significance of radiomics.
Keywords: biomarker; prognosis; radiomics; triple-negative breast cancer; tumor heterogeneity.
Copyright © 2022. Published by Elsevier Inc.
Conflict of interest statement
Declaration of interests L.J. is currently an employee of AstraZeneca.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

