Cell-free DNA 5-hydroxymethylcytosine is an emerging marker of acute myeloid leukemia
- PMID: 35859008
- PMCID: PMC9300744
- DOI: 10.1038/s41598-022-16685-3
Cell-free DNA 5-hydroxymethylcytosine is an emerging marker of acute myeloid leukemia
Abstract
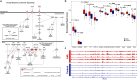
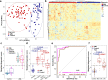
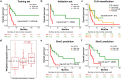
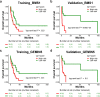
Aberrant changes in 5-hydroxymethylcytosine (5hmC) are a unique epigenetic feature in many cancers including acute myeloid leukemia (AML). However, genome-wide analysis of 5hmC in plasma cell-free DNA (cfDNA) remains unexploited in AML patients. We used a highly sensitive and robust nano-5hmC-Seal technology and profiled genome-wide 5hmC distribution in 239 plasma cfDNA samples from 103 AML patients and 81 non-cancer controls. We developed a 5hmC diagnostic model that precisely differentiates AML patients from controls with high sensitivity and specificity. We also developed a 5hmC prognostic model that accurately predicts prognosis in AML patients. High weighted prognostic scores (wp-scores) in AML patients were significantly associated with adverse overall survival (OS) in both training (P = 3.31e-05) and validation (P = 0.000464) sets. The wp-score was also significantly associated with genetic risk stratification and displayed dynamic changes with varied disease burden. Moreover, we found that high wp-scores in a single gene, BMS1 and GEMIN5 predicted OS in AML patients in both the training set (P = 0.023 and 0.031, respectively) and validation set (P = 9.66e-05 and 0.011, respectively). Lastly, our study demonstrated the genome-wide landscape of DNA hydroxymethylation in AML and revealed critical genes and pathways related to AML diagnosis and prognosis. Our data reveal plasma cfDNA 5hmC signatures as sensitive and accurate markers for AML diagnosis and prognosis. Plasma cfDNA 5hmC analysis will be an effective and minimally invasive tool for AML management.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Cell-Free DNA 5-Hydroxymethylcytosine Signatures for Lung Cancer Prognosis.Cells. 2024 Feb 6;13(4):298. doi: 10.3390/cells13040298. Cells. 2024. PMID: 38391911 Free PMC article.
-
Classification of Acute Myeloid Leukemia by Cell-Free DNA 5-Hydroxymethylcytosine.Genes (Basel). 2023 May 28;14(6):1180. doi: 10.3390/genes14061180. Genes (Basel). 2023. PMID: 37372359 Free PMC article.
-
Prognostic implications of 5-hydroxymethylcytosines from circulating cell-free DNA in diffuse large B-cell lymphoma.Blood Adv. 2019 Oct 8;3(19):2790-2799. doi: 10.1182/bloodadvances.2019000175. Blood Adv. 2019. PMID: 31570490 Free PMC article.
-
Cell-Free DNA Hydroxymethylation in Cancer: Current and Emerging Detection Methods and Clinical Applications.Genes (Basel). 2024 Sep 3;15(9):1160. doi: 10.3390/genes15091160. Genes (Basel). 2024. PMID: 39336751 Free PMC article. Review.
-
Towards precision medicine: advances in 5-hydroxymethylcytosine cancer biomarker discovery in liquid biopsy.Cancer Commun (Lond). 2019 Mar 29;39(1):12. doi: 10.1186/s40880-019-0356-x. Cancer Commun (Lond). 2019. PMID: 30922396 Free PMC article. Review.
Cited by
-
Cell-Free DNA 5-Hydroxymethylcytosine Signatures for Lung Cancer Prognosis.Cells. 2024 Feb 6;13(4):298. doi: 10.3390/cells13040298. Cells. 2024. PMID: 38391911 Free PMC article.
-
Deep5hmC: Predicting genome-wide 5-Hydroxymethylcytosine landscape via a multimodal deep learning model.bioRxiv [Preprint]. 2024 Mar 6:2024.03.04.583444. doi: 10.1101/2024.03.04.583444. bioRxiv. 2024. Update in: Bioinformatics. 2024 Sep 2;40(9):btae528. doi: 10.1093/bioinformatics/btae528. PMID: 38496575 Free PMC article. Updated. Preprint.
-
An Investigation into Cell-Free DNA in Different Common Cancers.Mol Biotechnol. 2024 Dec;66(12):3462-3474. doi: 10.1007/s12033-023-00976-9. Epub 2023 Dec 10. Mol Biotechnol. 2024. PMID: 38071680 Review.
-
Cell-free DNA 5-hydroxymethylcytosine is highly sensitive for MRD assessment in acute myeloid leukemia.Clin Epigenetics. 2023 Aug 24;15(1):134. doi: 10.1186/s13148-023-01547-0. Clin Epigenetics. 2023. PMID: 37620919 Free PMC article.
-
Deep5hmC: predicting genome-wide 5-hydroxymethylcytosine landscape via a multimodal deep learning model.Bioinformatics. 2024 Sep 2;40(9):btae528. doi: 10.1093/bioinformatics/btae528. Bioinformatics. 2024. PMID: 39196755 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

