A Functional Interaction Between Y674-R685 Region of the SARS-CoV-2 Spike Protein and the Human α7 Nicotinic Receptor
- PMID: 35859025
- PMCID: PMC9299415
- DOI: 10.1007/s12035-022-02947-8
A Functional Interaction Between Y674-R685 Region of the SARS-CoV-2 Spike Protein and the Human α7 Nicotinic Receptor
Abstract
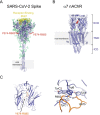
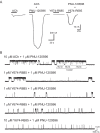
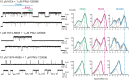
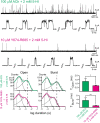
The α7 nicotinic acetylcholine receptor (nAChR) is present in neuronal and non-neuronal cells and has anti-inflammatory actions. Molecular dynamics simulations suggested that α7 nAChR interacts with a region of the SARS-CoV-2 spike protein (S), and a potential contribution of nAChRs to COVID-19 pathophysiology has been proposed. We applied whole-cell and single-channel recordings to determine whether a peptide corresponding to the Y674-R685 region of the S protein can directly affect α7 nAChR function. The S fragment exerts a dual effect on α7. It activates α7 nAChRs in the presence of positive allosteric modulators, in line with our previous molecular dynamics simulations showing favourable binding of this accessible region of the S protein to the nAChR agonist binding site. The S fragment also exerts a negative modulation of α7, which is evidenced by a profound concentration-dependent decrease in the durations of openings and activation episodes of potentiated channels and in the amplitude of macroscopic responses elicited by ACh. Our study identifies a potential functional interaction between α7 nAChR and a region of the S protein, thus providing molecular foundations for further exploring the involvement of nAChRs in COVID-19 pathophysiology.
Keywords: Neurotransmitter receptors; Nicotinic receptor; Patch-clamp; SARS-CoV-2 spike protein; Single-channel recordings.
© 2022. The Author(s), under exclusive licence to Springer Science+Business Media, LLC, part of Springer Nature.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
MeSH terms
Substances
Grants and funding
- PGI 24/B298/Universidad Nacional del Sur
- EP/M022609/1/Engineering and Physical Sciences Research Council
- BB/R016445/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- PICT 2017 1170/Agencia Nacional de Promoción Científica y Tecnológica
- BB/L01386X/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

