ADAR1 averts fatal type I interferon induction by ZBP1
- PMID: 35859176
- PMCID: PMC9329096
- DOI: 10.1038/s41586-022-04878-9
ADAR1 averts fatal type I interferon induction by ZBP1
Abstract
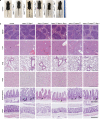
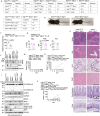
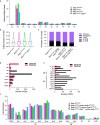
Mutations of the ADAR1 gene encoding an RNA deaminase cause severe diseases associated with chronic activation of type I interferon (IFN) responses, including Aicardi-Goutières syndrome and bilateral striatal necrosis1-3. The IFN-inducible p150 isoform of ADAR1 contains a Zα domain that recognizes RNA with an alternative left-handed double-helix structure, termed Z-RNA4,5. Hemizygous ADAR1 mutations in the Zα domain cause type I IFN-mediated pathologies in humans2,3 and mice6-8; however, it remains unclear how the interaction of ADAR1 with Z-RNA prevents IFN activation. Here we show that Z-DNA-binding protein 1 (ZBP1), the only other protein in mammals known to harbour Zα domains9, promotes type I IFN activation and fatal pathology in mice with impaired ADAR1 function. ZBP1 deficiency or mutation of its Zα domains reduced the expression of IFN-stimulated genes and largely prevented early postnatal lethality in mice with hemizygous expression of ADAR1 with mutated Zα domain (Adar1mZα/- mice). Adar1mZα/- mice showed upregulation and impaired editing of endogenous retroelement-derived complementary RNA reads, which represent a likely source of Z-RNAs activating ZBP1. Notably, ZBP1 promoted IFN activation and severe pathology in Adar1mZα/- mice in a manner independent of RIPK1, RIPK3, MLKL-mediated necroptosis and caspase-8-dependent apoptosis, suggesting a novel mechanism of action. Thus, ADAR1 prevents endogenous Z-RNA-dependent activation of pathogenic type I IFN responses by ZBP1, suggesting that ZBP1 could contribute to type I interferonopathies caused by ADAR1 mutations.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures











Comment in
-
ADAR1 inhibits ZBP1 activation by endogenous Z-RNA.Nat Rev Genet. 2022 Oct;23(10):581. doi: 10.1038/s41576-022-00525-1. Nat Rev Genet. 2022. PMID: 35922480 No abstract available.
-
Keeping immunostimulatory self-RNA under the rADAR.Nat Rev Rheumatol. 2022 Dec;18(12):681-682. doi: 10.1038/s41584-022-00859-4. Nat Rev Rheumatol. 2022. PMID: 36203062 No abstract available.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

