Time-keeping and decision-making in the cell cycle
- PMID: 35860005
- PMCID: PMC9184962
- DOI: 10.1098/rsfs.2021.0075
Time-keeping and decision-making in the cell cycle
Abstract
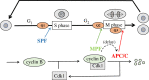
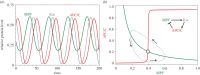
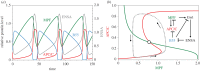
Cell growth, DNA replication, mitosis and division are the fundamental processes by which life is passed on from one generation of eukaryotic cells to the next. The eukaryotic cell cycle is intrinsically a periodic process but not so much a 'clock' as a 'copy machine', making new daughter cells as warranted. Cells growing under ideal conditions divide with clock-like regularity; however, if they are challenged with DNA-damaging agents or mitotic spindle disrupters, they will not progress to the next stage of the cycle until the damage is repaired. These 'decisions' (to exit and re-enter the cell cycle) are essential to maintain the integrity of the genome from generation to generation. A crucial challenge for molecular cell biologists in the 1990s was to unravel the genetic and biochemical mechanisms of cell cycle control in eukaryotes. Central to this effort were biochemical studies of the clock-like regulation of 'mitosis promoting factor' during synchronous mitotic cycles of fertilized frog eggs and genetic studies of the switch-like regulation of 'cyclin-dependent kinases' in yeast cells. In this review, we uncover some secrets of cell cycle regulation by mathematical modelling of increasingly more complex molecular regulatory networks of cell cycle 'clocks' and 'switches'.
Keywords: bistable switches; cell cycle checkpoints; cell cycle regulation; cyclin-dependent kinases; limit cycles; mathematical models.
© 2022 The Authors.
Conflict of interest statement
We declare we have no competing interests.
Figures




References
-
- Morgan DO. 2007. The cell cycle: principles of control. London, UK: New Science Press.
Publication types
Associated data
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

