Comparison of Rapid Cytokine Immunoassays for Functional Immune Phenotyping
- PMID: 35860253
- PMCID: PMC9289684
- DOI: 10.3389/fimmu.2022.940030
Comparison of Rapid Cytokine Immunoassays for Functional Immune Phenotyping
Abstract
Background: Cell-based functional immune-assays may allow for risk stratification of patients with complex, heterogeneous immune disorders such as sepsis. Given the heterogeneity of patient responses and the uncertain immune pathogenesis of sepsis, these assays must first be defined and calibrated in the healthy population.
Objective: Our objective was to compare the internal consistency and practicality of two immune assays that may provide data on surrogate markers of the innate and adaptive immune response. We hypothesized that a rapid turnaround, microfluidic-based immune assay (ELLA) would be comparable to a dual-color, enzyme-linked immunospot (ELISpot) assay in identifying tumor necrosis factor (TNF) and interferon (IFN)γ production following ex vivo whole blood stimulation.
Design: This was a prospective, observational cohort analysis. Whole blood samples from ten healthy, immune-competent volunteers were stimulated for either 4 hours or 18 hours with lipopolysaccharide, anti-CD3/anti-CD28 antibodies, or phorbol 12-myristate 13-acetate with ionomycin to interrogate innate and adaptive immune responses, respectively.
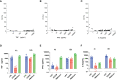
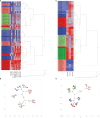
Measurements and main results: ELLA analysis produced more precise measurement of TNF and IFNγ concentrations as compared with ELISpot, as well as a four- to five-log10 dynamic range for TNF and IFNγ concentrations, as compared with a two-log10 dynamic range with ELISpot. Unsupervised clustering accurately predicted the ex vivo immune stimulant used for 90% of samples analyzed via ELLA, as compared with 72% of samples analyzed via ELISpot.
Conclusions: We describe, for the first time, a rapid and precise assay for functional interrogation of the innate and adaptive arms of the immune system in healthy volunteers. The advantages of the ELLA microfluidic platform may represent a step forward in generating a point-of-care test with clinical utility, for identifying deranged immune phenotypes in septic patients.
Keywords: cytokine; immune phenotype; immunoassay; interferon gamma; tumor necrosis factor.
Copyright © 2022 Bonavia, Samuelsen, Chroneos and Halstead.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Jacobs L, Berrens Z, Stenson EK, Zackoff MW, Danziger LA, Lahni P, et al. The Pediatric Sepsis Biomarker Risk Model (Persevere) Biomarkers Predict Clinical Deterioration and Mortality in Immunocompromised Children Evaluated for Infection. Sci Rep (2019) 9(1):424. doi: 10.1038/s41598-018-36743-z - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

