Comparative genomic analysis reveals new evidence of genus boundary for family Iridoviridae and explores qualified hallmark genes
- PMID: 35860404
- PMCID: PMC9284377
- DOI: 10.1016/j.csbj.2022.06.049
Comparative genomic analysis reveals new evidence of genus boundary for family Iridoviridae and explores qualified hallmark genes
Abstract
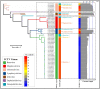
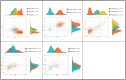
Members of the family Iridoviridae (iridovirids) are globally distributed and trigger adverse economic and ecological impacts on aquaculture and wildlife. Iridovirids taxonomy has previously been studied based on a limited number of genomes, but this is not suitable for the current and future virological studies as more iridovirids are emerging. In our study, 57 representative iridovirids genomes were selected from a total of 179 whole genomes available on NCBI. Then 18 core genes were screened out for members of the family Iridoviridae. Average amino acid sequence identity (AAI) analysis indicated that a cut-off value of 70% is more suitable for the current iridovirids genome database than ICTV-defined 50% threshold to better clarify viral genus boundaries. In addition, more subgroups were divided at genus level with the AAI threshold of 70%. This observation was further confirmed by genomic synteny analysis, codon usage preference analysis, genome GC content and length analysis, and phylogenic analysis. According to the pairwise comparison analysis of core genes, 9 hallmark genes were screened out to conduct preliminary identification and investigation at the genus level of iridovirids in a more convenient and economical manner.
Keywords: Codon usage; Core genes; Iridoviridae; Phylogenetics; Synteny analysis; Taxonomy.
© 2022 The Author(s).
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures







Similar articles
-
The Insights of Genomic Synteny and Codon Usage Preference on Genera Demarcation of Iridoviridae Family.Front Microbiol. 2021 Mar 31;12:657887. doi: 10.3389/fmicb.2021.657887. eCollection 2021. Front Microbiol. 2021. PMID: 33868215 Free PMC article.
-
Molecular and Ecological Studies of a Virus Family (Iridoviridae) Infecting Invertebrates and Ectothermic Vertebrates.Viruses. 2019 Jun 9;11(6):538. doi: 10.3390/v11060538. Viruses. 2019. PMID: 31181817 Free PMC article.
-
Genomic analysis of Poxviridae and exploring qualified gene sequences for phylogenetics.Comput Struct Biotechnol J. 2021 Sep 28;19:5479-5486. doi: 10.1016/j.csbj.2021.09.031. eCollection 2021. Comput Struct Biotechnol J. 2021. PMID: 34712393 Free PMC article.
-
Ranaviruses and other members of the family Iridoviridae: Their place in the virosphere.Virology. 2017 Nov;511:259-271. doi: 10.1016/j.virol.2017.06.007. Epub 2017 Jun 23. Virology. 2017. PMID: 28648249 Review.
-
Invertebrate Iridoviruses: A Glance over the Last Decade.Viruses. 2018 Mar 30;10(4):161. doi: 10.3390/v10040161. Viruses. 2018. PMID: 29601483 Free PMC article. Review.
Cited by
-
VIGA: a one-stop tool for eukaryotic virus identification and genome assembly from next-generation-sequencing data.Brief Bioinform. 2023 Nov 22;25(1):bbad444. doi: 10.1093/bib/bbad444. Brief Bioinform. 2023. PMID: 38048079 Free PMC article.
-
Molecular mechanism of the interaction between Megalocytivirus-induced virus-mock basement membrane (VMBM) and lymphatic endothelial cells.J Virol. 2023 Nov 30;97(11):e0048023. doi: 10.1128/jvi.00480-23. Epub 2023 Oct 25. J Virol. 2023. PMID: 37877715 Free PMC article.
-
Codon usage bias analysis of the gene encoding NAD+-dependent DNA ligase protein of Invertebrate iridescent virus 6.Arch Microbiol. 2023 Oct 9;205(11):352. doi: 10.1007/s00203-023-03688-5. Arch Microbiol. 2023. PMID: 37812231
-
Comparative Analyses of Bacteriophage Genomes.Methods Mol Biol. 2024;2802:427-453. doi: 10.1007/978-1-0716-3838-5_14. Methods Mol Biol. 2024. PMID: 38819567
-
Viral genomic methylation and the interspecies evolutionary relationships of ranavirus.PLoS Pathog. 2024 Nov 25;20(11):e1012736. doi: 10.1371/journal.ppat.1012736. eCollection 2024 Nov. PLoS Pathog. 2024. PMID: 39585924 Free PMC article.
References
-
- Gray M.J., Chinchar V.G. Ranaviruses: lethal pathogens of ectothermic vertebrates. Springer. Nature. 2015
-
- Zhao R., Geng Y., Qin Z., Wang K., Ouyang P., Chen D., et al. A new ranavirus of the Santee-Cooper group invades largemouth bass (Micropterus salmoides) culture in southwest China. Aquaculture. 2020;526
LinkOut - more resources
Full Text Sources
Miscellaneous
