The Cellular and Molecular Landscape of Synchronous Pediatric Sialoblastoma and Hepatoblastoma
- PMID: 35860547
- PMCID: PMC9289541
- DOI: 10.3389/fonc.2022.893206
The Cellular and Molecular Landscape of Synchronous Pediatric Sialoblastoma and Hepatoblastoma
Abstract
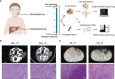
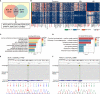
Sialoblastoma (SBL) is an infrequent embryonal malignant tumor originating from the salivary gland, resembling primitive salivary gland anlage, whereas hepatoblastoma (HB) is the most common pediatric liver malignancy. The simultaneous occurrence of both tumors is extremely rare. Here we reported a case of a 6-month-old infant diagnosed with synchronous SBL and HB. The patient received neoadjuvant chemotherapy followed by surgical resection. Fresh tissues of both tumors were collected before and after chemotherapy, which were further profiled by whole exome sequencing (WES) and single-cell RNA sequencing (scRNA-seq). WES analysis revealed potential somatic driver mutation PIK3CA p.Glu454Lys for SBL and canonical mutation CTNNB1 p.Ser45Pro for HB. No shared somatic variants or common copy number alterations were found between SBL and HB primary tumor samples. Though scRNA-seq, single-cell atlases were constructed for both tumors. SBL may recapitulate a pre-acinar stage in the development of salivary gland, including basaloid, duct-like, myoepithelial-like, and cycling phenotypes. In the meantime, HB was composed of tumor cells resembling different stages of the liver, including hepatocyte-like, hepatic progenitor-like, and hepatoblast-like cells. After chemotherapy, both tumors were induced into a more mature phenotype. In terms of transcriptional signatures, SBL and HB showed enhanced expression of epithelial markers KRT8, KRT18, and essential embryo development genes SDC1, MDK, indicating the disruption of normal embryo epithelium development. Finally, heterozygous deleterious germline mutation BLM and FANCI were identified which could predispose the patient to higher cancer risk. It partially explained the reason for the co-occurrence of SBL and HB. Taken together, we provided valuable resources for deciphering cellular heterogeneity and adaptive change of tumor cells after chemotherapy for synchronous SBL and HB, providing insights into the mechanisms leading to synchronous pediatric tumors.
Keywords: embryonal neoplasm; genetic predisposition to cancer; hepatoblastoma; sialoblastoma; single cell analysis.
Copyright © 2022 Yang, Zhan, Li, Dai, He, Ye, Meng, Chen, Dong, Chen, Chen, Dong, Li, Zheng, Li, Yao and Dong.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

