Counting Degrons: Lessons From Multivalent Substrates for Targeted Protein Degradation
- PMID: 35860655
- PMCID: PMC9289945
- DOI: 10.3389/fphys.2022.913063
Counting Degrons: Lessons From Multivalent Substrates for Targeted Protein Degradation
Abstract
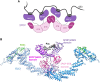
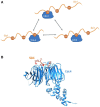
E3s comprise a structurally diverse group of at least 800 members, most of which target multiple substrates through specific and regulated protein-protein interactions. These interactions typically rely on short linear motifs (SLiMs), called "degrons", in an intrinsically disordered region (IDR) of the substrate, with variable rules of engagement governing different E3-docking events. These rules of engagement are of importance to the field of targeted protein degradation (TPD), where substrate ubiquitination and destruction require tools to effectively harness ubiquitin ligases (E3s). Substrates are often found to contain multiple degrons, or multiple copies of a degron, contributing to the affinity and selectivity of the substrate for its E3. One important paradigm for E3-substrate docking is presented by the Anaphase-Promoting Complex/Cyclosome (APC/C), a multi-subunit E3 ligase that targets hundreds of proteins for destruction during mitotic exit. APC/C substrate targeting takes place in an ordered manner thought to depend on tightly regulated interactions of substrates, with docking sites provided by the substoichiometric APC/C substrate adaptors and coactivators, Cdc20 or Cdh1/FZR1. Both structural and functional studies of individual APC/C substrates indicate that productive ubiquitination usually requires more than one degron, and that degrons are of different types docking to distinct sites on the coactivators. However, the dynamic nature of APC/C substrate recruitment, and the influence of multiple degrons, remains poorly understood. Here we review the significance of multiple degrons in a number of E3-substrate interactions that have been studied in detail, illustrating distinct kinetic effects of multivalency and allovalency, before addressing the role of multiple degrons in APC/C substrates, key to understanding ordered substrate destruction by APC/C. Lastly, we consider how lessons learnt from these studies can be applied in the design of TPD tools.
Keywords: E3-substrate interaction; SLiM; degron; multivalency; targeted protein degradation; ubiquitin ligase.
Copyright © 2022 Okoye, Rowling, Itzhaki and Lindon.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Building a Regulatory Network with Short Linear Sequence Motifs: Lessons from the Degrons of the Anaphase-Promoting Complex.Mol Cell. 2016 Oct 6;64(1):12-23. doi: 10.1016/j.molcel.2016.09.006. Mol Cell. 2016. PMID: 27716480 Free PMC article. Review.
-
Ubiquitination site preferences in anaphase promoting complex/cyclosome (APC/C) substrates.Open Biol. 2013 Sep 4;3(9):130097. doi: 10.1098/rsob.130097. Open Biol. 2013. PMID: 24004664 Free PMC article.
-
Activation of the APC/C ubiquitin ligase by enhanced E2 efficiency.Curr Biol. 2014 Jul 7;24(13):1556-62. doi: 10.1016/j.cub.2014.05.052. Epub 2014 Jun 12. Curr Biol. 2014. PMID: 24930963 Free PMC article.
-
The pseudosubstrate inhibitor Acm1 inhibits the anaphase-promoting complex/cyclosome by combining high-affinity activator binding with disruption of Doc1/Apc10 function.J Biol Chem. 2019 Nov 15;294(46):17249-17261. doi: 10.1074/jbc.RA119.009468. Epub 2019 Sep 27. J Biol Chem. 2019. PMID: 31562243 Free PMC article.
-
Design Principles Involving Protein Disorder Facilitate Specific Substrate Selection and Degradation by the Ubiquitin-Proteasome System.J Biol Chem. 2016 Mar 25;291(13):6723-31. doi: 10.1074/jbc.R115.692665. Epub 2016 Feb 5. J Biol Chem. 2016. PMID: 26851277 Free PMC article. Review.
Cited by
-
A conditional strategy for cell-type-specific labeling of endogenous excitatory synapses in Drosophila.Cell Rep Methods. 2023 May 11;3(5):100477. doi: 10.1016/j.crmeth.2023.100477. eCollection 2023 May 22. Cell Rep Methods. 2023. PMID: 37323572 Free PMC article.
-
Recognition of small Tim chaperones by the mitochondrial Yme1 protease.bioRxiv [Preprint]. 2025 Jul 24:2025.07.23.666395. doi: 10.1101/2025.07.23.666395. bioRxiv. 2025. PMID: 40777366 Free PMC article. Preprint.
-
Controlling CRISPR-Cas9 genome editing in human cells using a molecular glue degrader.Mol Ther Nucleic Acids. 2025 Jul 21;36(3):102640. doi: 10.1016/j.omtn.2025.102640. eCollection 2025 Sep 9. Mol Ther Nucleic Acids. 2025. PMID: 40799508 Free PMC article.
-
Overexpression of a Novel Noxo1 Mutant Increases Ros Production and Noxo1 Relocalisation.Int J Mol Sci. 2023 Feb 28;24(5):4663. doi: 10.3390/ijms24054663. Int J Mol Sci. 2023. PMID: 36902094 Free PMC article.
-
Investigating the Interactions of the Cucumber Mosaic Virus 2b Protein with the Viral 1a Replicase Component and the Cellular RNA Silencing Factor Argonaute 1.Viruses. 2024 Apr 25;16(5):676. doi: 10.3390/v16050676. Viruses. 2024. PMID: 38793558 Free PMC article.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

