Advances and Trends in Omics Technology Development
- PMID: 35860739
- PMCID: PMC9289742
- DOI: 10.3389/fmed.2022.911861
Advances and Trends in Omics Technology Development
Abstract
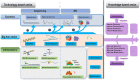
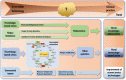
The human history has witnessed the rapid development of technologies such as high-throughput sequencing and mass spectrometry that led to the concept of "omics" and methodological advancement in systematically interrogating a cellular system. Yet, the ever-growing types of molecules and regulatory mechanisms being discovered have been persistently transforming our understandings on the cellular machinery. This renders cell omics seemingly, like the universe, expand with no limit and our goal toward the complete harness of the cellular system merely impossible. Therefore, it is imperative to review what has been done and is being done to predict what can be done toward the translation of omics information to disease control with minimal cell perturbation. With a focus on the "four big omics," i.e., genomics, transcriptomics, proteomics, metabolomics, we delineate hierarchies of these omics together with their epiomics and interactomics, and review technologies developed for interrogation. We predict, among others, redoxomics as an emerging omics layer that views cell decision toward the physiological or pathological state as a fine-tuned redox balance.
Keywords: mass spectrometry; next generation sequencing; omics; redoxomics; third generation sequencing.
Copyright © 2022 Dai and Shen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
Publication types
LinkOut - more resources
Full Text Sources

