A secreted fungal subtilase interferes with rice immunity via degradation of SUPPRESSOR OF G2 ALLELE OF skp1
- PMID: 35861434
- PMCID: PMC9516721
- DOI: 10.1093/plphys/kiac334
A secreted fungal subtilase interferes with rice immunity via degradation of SUPPRESSOR OF G2 ALLELE OF skp1
Abstract
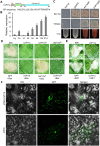
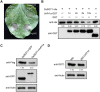
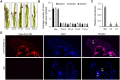
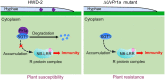
Serine protease subtilase, found widely in both eukaryotes and prokaryotes, participates in various biological processes. However, how fungal subtilase regulates plant immunity is a major concern. Here, we identified a secreted fungal subtilase, UvPr1a, from the rice false smut (RFS) fungus Ustilaginoidea virens. We characterized UvPr1a as a virulence effector localized to the plant cytoplasm that inhibits plant cell death induced by Bax. Heterologous expression of UvPr1a in rice (Oryza sativa) enhanced plant susceptibility to rice pathogens. UvPr1a interacted with the important rice protein SUPPRESSOR OF G2 ALLELE OF skp1 (OsSGT1), a positive regulator of innate immunity against multiple rice pathogens, degrading OsSGT1 in a protease activity-dependent manner. Furthermore, host-induced gene silencing of UvPr1a compromised disease resistance of rice plants. Our work reveals a previously uncharacterized fungal virulence strategy in which a fungal pathogen secretes a subtilase to interfere with rice immunity through degradation of OsSGT1, thereby promoting infection. These genetic resources provide tools for introducing RFS resistance and further our understanding of plant-pathogen interactions.
© American Society of Plant Biologists 2022. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures




Similar articles
-
Ustilaginoidea virens, an emerging pathogen of rice: the dynamic interplay between the pathogen virulence strategies and host defense.Planta. 2024 Sep 11;260(4):92. doi: 10.1007/s00425-024-04523-x. Planta. 2024. PMID: 39261328 Review.
-
A secreted fungal laccase targets the receptor kinase OsSRF3 to inhibit OsBAK1-OsSRF3-mediated immunity in rice.Nat Commun. 2024 Sep 10;15(1):7891. doi: 10.1038/s41467-024-52204-w. Nat Commun. 2024. PMID: 39256395 Free PMC article.
-
The Conserved Effector UvHrip1 interacts with OsHGW, and Infection of Ustilaginoidea virens Regulates Defense- and Heading Date-Related Signaling Pathway.Int J Mol Sci. 2020 May 10;21(9):3376. doi: 10.3390/ijms21093376. Int J Mol Sci. 2020. PMID: 32397668 Free PMC article.
-
SnRK1A-mediated phosphorylation of a cytosolic ATPase positively regulates rice innate immunity and is inhibited by Ustilaginoidea virens effector SCRE1.New Phytol. 2022 Nov;236(4):1422-1440. doi: 10.1111/nph.18460. Epub 2022 Sep 17. New Phytol. 2022. PMID: 36068953
-
Ustilaginoidea virens: Insights into an Emerging Rice Pathogen.Annu Rev Phytopathol. 2020 Aug 25;58:363-385. doi: 10.1146/annurev-phyto-010820-012908. Epub 2020 May 4. Annu Rev Phytopathol. 2020. PMID: 32364825 Review.
Cited by
-
Ustilaginoidea virens-secreted effector Uv1809 suppresses rice immunity by enhancing OsSRT2-mediated histone deacetylation.Plant Biotechnol J. 2024 Jan;22(1):148-164. doi: 10.1111/pbi.14174. Epub 2023 Sep 16. Plant Biotechnol J. 2024. PMID: 37715970 Free PMC article.
-
ThSCSP_12: Novel Effector in Tilletia horrida That Induces Cell Death and Defense Responses in Non-Host Plants.Int J Mol Sci. 2022 Nov 25;23(23):14752. doi: 10.3390/ijms232314752. Int J Mol Sci. 2022. PMID: 36499087 Free PMC article.
-
Regulation of plant immunity through histone H3 β-hydroxybutyrylation-mediated transcriptional control.Nat Commun. 2025 Jul 17;16(1):6588. doi: 10.1038/s41467-025-61474-x. Nat Commun. 2025. PMID: 40675960 Free PMC article.
-
Ustilaginoidea virens, an emerging pathogen of rice: the dynamic interplay between the pathogen virulence strategies and host defense.Planta. 2024 Sep 11;260(4):92. doi: 10.1007/s00425-024-04523-x. Planta. 2024. PMID: 39261328 Review.
-
A secreted fungal laccase targets the receptor kinase OsSRF3 to inhibit OsBAK1-OsSRF3-mediated immunity in rice.Nat Commun. 2024 Sep 10;15(1):7891. doi: 10.1038/s41467-024-52204-w. Nat Commun. 2024. PMID: 39256395 Free PMC article.
References
-
- Azevedo C, Sadanandom A, Kitagawa K, Freialdenhoven A, Shirasu K, Schulze-Lefert P (2002) The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science 295: 2073–2076 - PubMed
-
- Baker D, Shiau AK, Agard DA (1993) The role of pro regions in protein folding. Curr Opin Cell Biol 5: 966–970 - PubMed
-
- Bryan PN (2002) Prodomains and protein folding catalysis. Chem Rev 102: 4805–4816 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

