Time Series Genomics of Pseudomonas aeruginosa Reveals the Emergence of a Hypermutator Phenotype and Within-Host Evolution in Clinical Inpatients
- PMID: 35861512
- PMCID: PMC9430856
- DOI: 10.1128/spectrum.00057-22
Time Series Genomics of Pseudomonas aeruginosa Reveals the Emergence of a Hypermutator Phenotype and Within-Host Evolution in Clinical Inpatients
Abstract
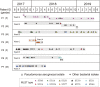
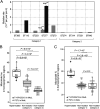
Pseudomonas aeruginosa, a common opportunistic pathogen, is one of the leading etiological agents of nosocomial infections. Many previous studies have reported the nosocomial transmission and epidemiology of P. aeruginosa infections. However, longitudinal studies regarding the dynamics of P. aeruginosa colonization and infection in health care settings are limited. We obtained longitudinal samples from aged patients with prolonged intensive care unit (ICU) stays (~4 to 19 months). P. aeruginosa was isolated from 71 samples obtained from seven patients and characterized by whole-genome sequencing. The P. aeruginosa isolates were assigned to 10 clonal complexes, and turnover of main clones was observed in sequential sputum samples from two patients. By comparing intraclonal genomic diversities, we identified two clones that had significantly higher numbers of single nucleotide polymorphisms and variations in homopolymeric sequences than the other clones, indicating a hypermutator phenotype. These hypermutator clones were associated with mutations T147I/G521S and P27L in the MutL protein, and their mutation rates were estimated to be 3.20 × 10-5 and 6.59 × 10-5 per year per nucleotide, respectively. We also identified 24 recurrently mutated genes that exhibited intraclonal diversity in two or more clones. Notably, one recurrent mutation, S698F in FptA, was observed in four clones. These findings suggest that convergent microevolution and adaption of P. aeruginosa occur in long-term ICU patients. IMPORTANCE Pseudomonas aeruginosa is a predominant opportunistic pathogen that causes nosocomial infections. Inappropriate empirical therapy can lead to prolonged hospital stays and increased mortality. In our study of sequential P. aeruginosa isolates from inpatients, high intrahost diversity was observed, including switching of clones and the emergence of a hypermutator phenotype. Recurrently mutated genes also suggested that convergent microevolution and adaption of P. aeruginosa occur in inpatients, and genomic diversity is associated with differences in multiple-drug-resistance profiles. Taken together, our findings highlight the importance of longitudinal surveillance of nosocomial P. aeruginosa clones.
Keywords: Pseudomonas aeruginosa; hypermutator; longitudinal study; nosocomial infections; whole-genome sequencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Genomic and phenotypic inconsistencies in Pseudomonas aeruginosa resistome among intensive care patients.Front Cell Infect Microbiol. 2024 Jun 21;14:1335096. doi: 10.3389/fcimb.2024.1335096. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 38975326 Free PMC article.
-
Evolved Aztreonam Resistance Is Multifactorial and Can Produce Hypervirulence in Pseudomonas aeruginosa.mBio. 2017 Oct 31;8(5):e00517-17. doi: 10.1128/mBio.00517-17. mBio. 2017. PMID: 29089424 Free PMC article.
-
Hypermutator Pseudomonas aeruginosa Exploits Multiple Genetic Pathways To Develop Multidrug Resistance during Long-Term Infections in the Airways of Cystic Fibrosis Patients.Antimicrob Agents Chemother. 2020 Apr 21;64(5):e02142-19. doi: 10.1128/AAC.02142-19. Print 2020 Apr 21. Antimicrob Agents Chemother. 2020. PMID: 32071060 Free PMC article.
-
Host and Pathogen Biomarkers for Severe Pseudomonas aeruginosa Infections.J Infect Dis. 2017 Feb 15;215(suppl_1):S44-S51. doi: 10.1093/infdis/jiw299. J Infect Dis. 2017. PMID: 28375513 Review.
-
Antibiotic resistance in Pseudomonas aeruginosa - Mechanisms, epidemiology and evolution.Drug Resist Updat. 2019 May;44:100640. doi: 10.1016/j.drup.2019.07.002. Epub 2019 Jul 19. Drug Resist Updat. 2019. PMID: 31492517 Review.
Cited by
-
Comparison of Different Methods for Assaying the In Vitro Activity of Cefiderocol against Carbapenem-Resistant Pseudomonas aeruginosa Strains: Influence of Bacterial Inoculum.Antibiotics (Basel). 2024 Jul 18;13(7):663. doi: 10.3390/antibiotics13070663. Antibiotics (Basel). 2024. PMID: 39061345 Free PMC article.
-
Molecular epidemiology and genomic dynamics of Pseudomonas aeruginosa isolates causing relapse infections.Microbiol Spectr. 2023 Sep 28;11(5):e0531222. doi: 10.1128/spectrum.05312-22. Online ahead of print. Microbiol Spectr. 2023. PMID: 37768065 Free PMC article.
-
Phenotypes of a Pseudomonas aeruginosa hypermutator lineage that emerged during prolonged mechanical ventilation in a patient without cystic fibrosis.mSystems. 2024 Jan 23;9(1):e0048423. doi: 10.1128/msystems.00484-23. Epub 2023 Dec 22. mSystems. 2024. PMID: 38132670 Free PMC article.
References
-
- Cristina ML, Spagnolo AM, Orlando P, Perdelli F. 2013. The role of the environment in the spread of emerging pathogens in at-risk hospital wards. Rev Med Microbiol 24:104–112. doi:10.1097/MRM.0b013e328365c506. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

