PSMD12-Mediated M1 Ubiquitination of Influenza A Virus at K102 Regulates Viral Replication
- PMID: 35861516
- PMCID: PMC9364790
- DOI: 10.1128/jvi.00786-22
PSMD12-Mediated M1 Ubiquitination of Influenza A Virus at K102 Regulates Viral Replication
Abstract
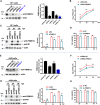
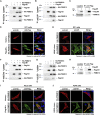
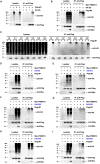
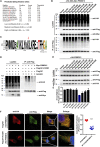
The M1 of influenza A virus (IAV) is important for the virus life cycle, especially for the assembly and budding of viruses, which is a multistep process that requires host factors. Identifying novel host proteins that interact with M1 and understanding their functions in IAV replication are of great interest in antiviral drug development. In this study, we identified 19 host proteins in DF1 cells suspected to interact with the M1 protein of an H5N6 virus through immunoprecipitation (IP)/mass spectrometry. Among them, PSMD12, a 26S proteasome regulatory subunit, was shown to interact with influenza M1, acting as a positive host factor in IAV replication in avian and human cells. The data showed that PSMD12 promoted K63-linked ubiquitination of M1 at the K102 site. H5N6 and PR8 with an M1-K102 site mutant displayed a significantly weaker replication ability than the wild-type viruses. Mechanistically, PSMD12 promoted M1-M2 virus-like particle (VLP) release, and an M1-K102 mutation disrupted the formation of supernatant M1-M2 VLPs. An H5N6 M1-K102 site mutation or knockdown PSMD12 disrupted the budding release of the virus in chicken embryo fibroblast (CEF) cells, which was confirmed by transmission electron microscopy. Further study confirmed that M1-K102 site mutation significantly affected the virulence of H5N6 and PR8 viruses in mice. In conclusion, we report the novel host factor PSMD12 which affects the replication of influenza virus by mediating K63-linked ubiquitination of M1 at K102. These findings provide novel insight into the interactions between IAV and host cells, while suggesting an important target for anti-influenza virus drug research. IMPORTANCE M1 is proposed to play multiple biologically important roles in the life cycle of IAV, which relies largely on host factors. This study is the first one to identify that PSMD12 interacts with M1, mediates K63-linked ubiquitination of M1 at the K102 site, and thus positively regulates influenza virus proliferation. PSMD12 promoted M1-M2 VLP egress, and an M1-K102 mutation affected the M1-M2 VLP formation. Furthermore, we demonstrate the importance of this site to the morphology and budding of influenza viruses by obtaining mutant viruses, and the M1 ubiquitination regulator PSMD12 has a similar function to the M1 K102 mutation in regulating virus release and virus morphology. Additionally, we confirm the reduced virulence of H5N6 and PR8 (H1N1) viruses carrying the M1-K102 site mutation in mice. These findings provide novel insights into IAV interactions with host cells and suggest a valid and highly conserved candidate target for antiviral drug development.
Keywords: K102 site; K63-linked ubiquitination; M1 protein; PSMD12; influenza A virus; replication.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Gong W, He X, Huang K, Zhang Y, Li C, Yang Y, Zou Z, Jin M. 2021. Interaction of nuclear export protein with G protein pathway suppressor 2 (GPS2) facilitates influenza A virus replication by weakening the inhibition of GPS2 to RNA synthesis and ribonucleoprotein assembly. J Virol 95:e00008-21. 10.1128/JVI.00008-21. - DOI - PMC - PubMed
-
- Watanabe T, Kawakami E, Shoemaker JE, Lopes TJS, Matsuoka Y, Tomita Y, Kozuka-Hata H, Gorai T, Kuwahara T, Takeda E, Nagata A, Takano R, Kiso M, Yamashita M, Sakai-Tagawa Y, Katsura H, Nonaka N, Fujii H, Fujii K, Sugita Y, Noda T, Goto H, Fukuyama S, Watanabe S, Neumann G, Oyama M, Kitano H, Kawaoka Y. 2014. Influenza virus-host interactome screen as a platform for antiviral drug development. Cell Host Microbe 16:795–805. 10.1016/j.chom.2014.11.002. - DOI - PMC - PubMed
-
- Gannage M, Dormann D, Albrecht R, Dengjel J, Torossi T, Rämer PC, Lee M, Strowig T, Arrey F, Conenello G, Pypaert M, Andersen J, García-Sastre A, Münz C. 2009. Matrix protein 2 of influenza A virus blocks autophagosome fusion with lysosomes. Cell Host Microbe 6:367–380. 10.1016/j.chom.2009.09.005. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

