Clock genes rescue nphp mutations in zebrafish
- PMID: 35861640
- PMCID: PMC9759334
- DOI: 10.1093/hmg/ddac160
Clock genes rescue nphp mutations in zebrafish
Abstract
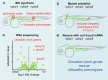
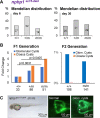
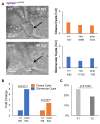
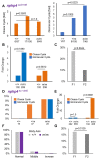
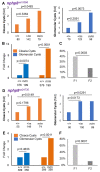
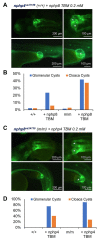
The zebrafish pronephros model, using morpholino oligonucleotides (MO) to deplete target genes, has been extensively used to characterize human ciliopathy phenotypes. Recently, discrepancies between MO and genetically defined mutants have questioned this approach. We analyzed zebrafish with mutations in the nphp1-4-8 module to determine the validity of MO-based results. While MO-mediated depletion resulted in glomerular cyst and cloaca malformation, these ciliopathy-typical manifestations were observed at a much lower frequency in zebrafish embryos with defined nphp mutations. All nphp1-4-8 mutant zebrafish were viable and displayed decreased manifestations in the next (F2) generation, lacking maternal RNA contribution. While genetic compensation was further supported by the observation that nphp4-deficient mutants became partially refractory to MO-based nphp4 depletion, zebrafish embryos, lacking one nphp gene, became more sensitive to MO-based depletion of additional nphp genes. Transcriptome analysis of nphp8 mutant embryos revealed an upregulation of the circadian clock genes cry1a and cry5. MO-mediated depletion of cry1a and cry5 caused ciliopathy phenotypes in wild-type embryos, while cry1a and cry5 depletion in maternal zygotic nphp8 mutant embryos increased the frequency of glomerular cysts compared to controls. Importantly, cry1a and cry5 rescued the nephropathy-related phenotypes in nphp1, nphp4 or nphp8-depleted zebrafish embryos. Our results reveal that nphp mutant zebrafish resemble the MO-based phenotypes, albeit at a much lower frequency. Rapid adaption through upregulation of circadian clock genes seems to ameliorate the loss of nphp genes, contributing to phenotypic differences.
© The Author(s) 2022. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures



Similar articles
-
Evolutionarily conserved genetic interactions between nphp-4 and bbs-5 mutations exacerbate ciliopathy phenotypes.Genetics. 2022 Jan 4;220(1):iyab209. doi: 10.1093/genetics/iyab209. Genetics. 2022. PMID: 34850872 Free PMC article.
-
Inversin (NPHP2) and Vangl2 are required for normal zebrafish cloaca formation.Biochem Biophys Res Commun. 2023 Sep 17;673:9-15. doi: 10.1016/j.bbrc.2023.06.058. Epub 2023 Jun 19. Biochem Biophys Res Commun. 2023. PMID: 37352572
-
Mutations of ADAMTS9 Cause Nephronophthisis-Related Ciliopathy.Am J Hum Genet. 2019 Jan 3;104(1):45-54. doi: 10.1016/j.ajhg.2018.11.003. Am J Hum Genet. 2019. PMID: 30609407 Free PMC article.
-
Nephronophthisis-associated ciliopathies.J Am Soc Nephrol. 2007 Jun;18(6):1855-71. doi: 10.1681/ASN.2006121344. Epub 2007 May 18. J Am Soc Nephrol. 2007. PMID: 17513324 Review.
-
Insights Gained From Zebrafish Models for the Ciliopathy Joubert Syndrome.Front Genet. 2022 Jun 30;13:939527. doi: 10.3389/fgene.2022.939527. eCollection 2022. Front Genet. 2022. PMID: 35846153 Free PMC article. Review.
Cited by
-
Targeting GLP-1 Signaling Ameliorates Cystogenesis in a Zebrafish Model of Nephronophthisis.Int J Mol Sci. 2025 Jul 30;26(15):7366. doi: 10.3390/ijms26157366. Int J Mol Sci. 2025. PMID: 40806500 Free PMC article.
References
-
- Otto, E., Kispert, A., Schätzle, S., Lescher, B., Rensing, C. and Hildebrandt, F. (2000) Nephrocystin: gene expression and sequence conservation between human, mouse, and Caenorhabditis elegans. JASN, 11, 270–282. - PubMed
-
- Wolf, M.T.F., Lee, J., Panther, F., Otto, E.A., Guan, K.-L. and Hildebrandt, F. (2005) Expression and phenotype analysis of the nephrocystin-1 and nephrocystin-4 homologs in Caenorhabditis elegans. JASN, 16, 676–687. - PubMed
-
- Jauregui, A.R. and Barr, M.M. (2005) Functional characterization of the C. elegans nephrocystins NPHP-1 and NPHP-4 and their role in cilia and male sensory behaviors. Exp. Cell Res., 305, 333–342. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

