Developing a Long COVID Phenotype for Postacute COVID-19 in a National Primary Care Sentinel Cohort: Observational Retrospective Database Analysis
- PMID: 35861678
- PMCID: PMC9374163
- DOI: 10.2196/36989
Developing a Long COVID Phenotype for Postacute COVID-19 in a National Primary Care Sentinel Cohort: Observational Retrospective Database Analysis
Abstract
Background: Following COVID-19, up to 40% of people have ongoing health problems, referred to as postacute COVID-19 or long COVID (LC). LC varies from a single persisting symptom to a complex multisystem disease. Research has flagged that this condition is underrecorded in primary care records, and seeks to better define its clinical characteristics and management. Phenotypes provide a standard method for case definition and identification from routine data and are usually machine-processable. An LC phenotype can underpin research into this condition.
Objective: This study aims to develop a phenotype for LC to inform the epidemiology and future research into this condition. We compared clinical symptoms in people with LC before and after their index infection, recorded from March 1, 2020, to April 1, 2021. We also compared people recorded as having acute infection with those with LC who were hospitalized and those who were not.
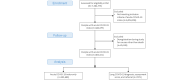
Methods: We used data from the Primary Care Sentinel Cohort (PCSC) of the Oxford Royal College of General Practitioners (RCGP) Research and Surveillance Centre (RSC) database. This network was recruited to be nationally representative of the English population. We developed an LC phenotype using our established 3-step ontological method: (1) ontological step (defining the reasoning process underpinning the phenotype, (2) coding step (exploring what clinical terms are available, and (3) logical extract model (testing performance). We created a version of this phenotype using Protégé in the ontology web language for BioPortal and using PhenoFlow. Next, we used the phenotype to compare people with LC (1) with regard to their symptoms in the year prior to acquiring COVID-19 and (2) with people with acute COVID-19. We also compared hospitalized people with LC with those not hospitalized. We compared sociodemographic details, comorbidities, and Office of National Statistics-defined LC symptoms between groups. We used descriptive statistics and logistic regression.
Results: The long-COVID phenotype differentiated people hospitalized with LC from people who were not and where no index infection was identified. The PCSC (N=7.4 million) includes 428,479 patients with acute COVID-19 diagnosis confirmed by a laboratory test and 10,772 patients with clinically diagnosed COVID-19. A total of 7471 (1.74%, 95% CI 1.70-1.78) people were coded as having LC, 1009 (13.5%, 95% CI 12.7-14.3) had a hospital admission related to acute COVID-19, and 6462 (86.5%, 95% CI 85.7-87.3) were not hospitalized, of whom 2728 (42.2%) had no COVID-19 index date recorded. In addition, 1009 (13.5%, 95% CI 12.73-14.28) people with LC were hospitalized compared to 17,993 (4.5%, 95% CI 4.48-4.61; P<.001) with uncomplicated COVID-19.
Conclusions: Our LC phenotype enables the identification of individuals with the condition in routine data sets, facilitating their comparison with unaffected people through retrospective research. This phenotype and study protocol to explore its face validity contributes to a better understanding of LC.
Keywords: BioPortal; COVID-19; SARS-CoV-2; Systematized Nomenclature of Medicine; biomedical ontologies; computerized; data accuracy; data extracts; digital tool; disease management; electronic health record; epidemiology; ethnicity; general practitioners; hospitalization; long COVID; medical record systems; phenotype; postacute COVID-19 syndrome; public health; social class; surveillance.
©Nikhil Mayor, Bernardo Meza-Torres, Cecilia Okusi, Gayathri Delanerolle, Martin Chapman, Wenjuan Wang, Sneha Anand, Michael Feher, Jack Macartney, Rachel Byford, Mark Joy, Piers Gatenby, Vasa Curcin, Trisha Greenhalgh, Brendan Delaney, Simon de Lusignan. Originally published in JMIR Public Health and Surveillance (https://publichealth.jmir.org), 11.08.2022.
Conflict of interest statement
Conflicts of Interest: SdL reports that through his university, he had grants from AstraZeneca, GlaxoSmithKlein (GSK), Sanofi, Seqirus, and Takeda for vaccine-related research and membership of advisory boards for AstraZeneca, Sanofi, and Seqirus. TG is a member of Independent SAGE. All other authors declare no conflicts of interest.
Figures






References
-
- Ayoubkhani D, King S, Bosworth M. Prevalence of Ongoing Symptoms Following Coronavirus (COVID-19) Infection in the UK: 2 December 2021. [2022-08-06]. https://tinyurl.com/4wbnxtrn .
-
- Deer RR, Rock MA, Vasilevsky N, Carmody L, Rando H, Anzalone AJ, Basson MD, Bennett TD, Bergquist T, Boudreau EA, Bramante CT, Byrd JB, Callahan TJ, Chan LE, Chu H, Chute CG, Coleman BD, Davis HE, Gagnier J, Greene CS, Hillegass WB, Kavuluru R, Kimble WD, Koraishy FM, Köhler S, Liang C, Liu F, Liu H, Madhira V, Madlock-Brown CR, Matentzoglu N, Mazzotti DR, McMurry JA, McNair DS, Moffitt RA, Monteith TS, Parker AM, Perry MA, Pfaff E, Reese JT, Saltz J, Schuff RA, Solomonides AE, Solway J, Spratt H, Stein GS, Sule AA, Topaloglu U, Vavougios GD, Wang L, Haendel MA, Robinson PN. Characterizing long COVID: deep phenotype of a complex condition. EBioMedicine. 2021 Dec;74:103722. doi: 10.1016/j.ebiom.2021.103722. https://linkinghub.elsevier.com/retrieve/pii/S2352-3964(21)00516-8 S2352-3964(21)00516-8 - DOI - PMC - PubMed
-
- Aiyegbusi OL, Hughes SE, Turner G, Rivera SC, McMullan C, Chandan JS, Haroon S, Price G, Davies EH, Nirantharakumar K, Sapey E, Calvert MJ, Study Group Tlc. Symptoms, complications and management of long COVID: a review. J R Soc Med. 2021 Sep 15;114(9):428–442. doi: 10.1177/01410768211032850. https://journals.sagepub.com/doi/10.1177/01410768211032850?url_ver=Z39.8... - DOI - DOI - PMC - PubMed
-
- Sudre CH, Murray B, Varsavsky T, Graham MS, Penfold RS, Bowyer RC, Pujol JC, Klaser K, Antonelli M, Canas LS, Molteni E, Modat M, Jorge Cardoso M, May A, Ganesh S, Davies R, Nguyen LH, Drew DA, Astley CM, Joshi AD, Merino J, Tsereteli N, Fall T, Gomez MF, Duncan EL, Menni C, Williams FMK, Franks PW, Chan AT, Wolf J, Ourselin S, Spector T, Steves CJ. Attributes and predictors of long COVID. Nat Med. 2021 Apr;27(4):626–631. doi: 10.1038/s41591-021-01292-y. https://europepmc.org/abstract/MED/33692530 10.1038/s41591-021-01292-y - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

