Inhibition of Epidermal Growth Factor Receptor Signaling by Antisense Oligonucleotides as a Novel Approach to Epidermal Growth Factor Receptor Inhibition
- PMID: 35861718
- PMCID: PMC9595651
- DOI: 10.1089/nat.2021.0101
Inhibition of Epidermal Growth Factor Receptor Signaling by Antisense Oligonucleotides as a Novel Approach to Epidermal Growth Factor Receptor Inhibition
Abstract
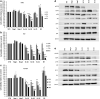
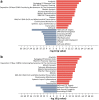
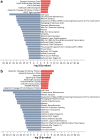
We report a novel method to inhibit epidermal growth factor receptor (EGFR) signaling using custom morpholino antisense oligonucleotides (ASOs) to drive expression of dominant negative mRNA isoforms of EGFR by ASO-induced exon skipping within the transmembrane (16) or tyrosine kinase domains (18 and 21). In vivo ASO formulations induced >95% exon skipping in several models of nonsmall cell lung cancer (NSCLC) and were comparable in efficacy to erlotinib in reducing colony formation, cell viability, and migration in EGFR mutant NSCLC (PC9). However, unlike erlotinib, ASOs maintained their efficacy in both erlotinib-resistant subclones (PC9-GR) and wild-type overexpressing EGFR models (H292), in which erlotinib had no significant effect. The most dramatic ASO-induced phenotype resulted from targeting the EGFR kinase domain directly, which resulted in maximal inhibition of phosphorylation of EGFR, Akt, and Erk in both PC9 and PC9GR cells. Phosphoproteomic mass spectrometry confirmed highly congruent impacts of exon 16-, 18-, and 21-directed ASOs compared with erlotinib on PC9 genome-wide cell signaling. Furthermore, EGFR-directed ASOs had no impact in EGFR-independent NSCLC models, confirming an EGFR-specific therapeutic mechanism. Further exploration of synergy of ASOs with existing tyrosine kinase inhibitors may offer novel clinical models to improve EGFR-targeted therapies for both mutant and wild-type NSCLC patients.
Keywords: antisense oligonucleotides; epidermal growth factor receptor; exon skipping; receptor tyrosine kinase; tyrosine kinase inhibitors.
Conflict of interest statement
T.R. holds a patent related to the therapeutic use of antisense oligonucleotides.
Figures


References
-
- Manning G, Whyte DB, Martinez R, Hunter T and Sudarsanam S. (2002). The protein kinase complement of the human genome. Science 298:1912–1934. - PubMed
-
- Blume-Jensen P and Hunter T. (2001). Oncogenic kinase signalling. Nature 411:355–365. - PubMed
-
- Hubbard SR. (1999). Structural analysis of receptor tyrosine kinases. Prog Biophys Mol Bio 71:343–358. - PubMed
-
- Yarden Y and Pines G. (2012). The ERBB network: at last, cancer therapy meets systems biology. Nat Rev Cancer 12:553–563. - PubMed
-
- Li SQ, Schmitz KR, Jeffrey PD, Wiltzius JJW, Kussie P and Ferguson KM. (2005). Structural basis for inhibition of the epidermal growth factor receptor by cetuximab. Cancer Cell 7:301–311. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

