Centriole growth is limited by the Cdk/Cyclin-dependent phosphorylation of Ana2/STIL
- PMID: 35861803
- PMCID: PMC9442473
- DOI: 10.1083/jcb.202205058
Centriole growth is limited by the Cdk/Cyclin-dependent phosphorylation of Ana2/STIL
Abstract
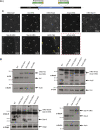
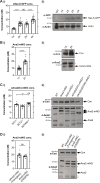
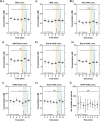
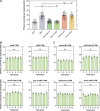
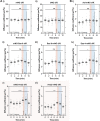
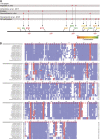
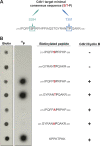
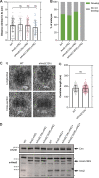
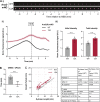
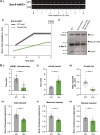
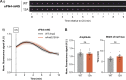
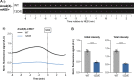
Centrioles duplicate once per cell cycle, but it is unclear how daughter centrioles assemble at the right time and place and grow to the right size. Here, we show that in Drosophila embryos the cytoplasmic concentrations of the key centriole assembly proteins Asl, Plk4, Ana2, Sas-6, and Sas-4 are low, but remain constant throughout the assembly process-indicating that none of them are limiting for centriole assembly. The cytoplasmic diffusion rate of Ana2/STIL, however, increased significantly toward the end of S-phase as Cdk/Cyclin activity in the embryo increased. A mutant form of Ana2 that cannot be phosphorylated by Cdk/Cyclins did not exhibit this diffusion change and allowed daughter centrioles to grow for an extended period. Thus, the Cdk/Cyclin-dependent phosphorylation of Ana2 seems to reduce the efficiency of daughter centriole assembly toward the end of S-phase. This helps to ensure that daughter centrioles stop growing at the correct time, and presumably also helps to explain why centrioles cannot duplicate during mitosis.
© 2022 Steinacker et al.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

