Insights into male androgenetic alopecia using comparative transcriptome profiling: hypoxia-inducible factor-1 and Wnt/β-catenin signalling pathways
- PMID: 35862273
- PMCID: PMC10087000
- DOI: 10.1111/bjd.21783
Insights into male androgenetic alopecia using comparative transcriptome profiling: hypoxia-inducible factor-1 and Wnt/β-catenin signalling pathways
Abstract
Background: The key pathophysiological changes in androgenetic alopecia (AGA) are limited to hair follicles (HFs) in frontal and vertex regions, sparing the occipital region.
Objectives: To identify biological differences among HF subpopulations.
Methods: Paired vertex and occipital HFs from 10 male donors with AGA were collected for RNA sequencing assay. Furthermore, HF and cell experiments were conducted on the identified key genes to reveal their roles in AGA.
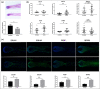
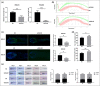
Results: Transcriptome profiles revealed that 506 mRNAs, 55 microRNAs and 127 long noncoding RNAs were differentially expressed in the AGA vertex HFs. Pathway analysis of mRNAs and microRNAs revealed involvement of the hypoxia-inducible factor (HIF)-1, Wnt/β-catenin, and focal adhesion pathways. Differential expression of HIF-1 prolyl hydroxylase enzymes (EGLN1, EGLN3) and Wnt/β-catenin pathway inhibitors (SERPINF1, SFRP2) was experimentally validated. In vitro studies revealed that reduction of EGLN1, EGLN3, SERPINF1 and SFRP2 stimulated proliferation of dermal papilla cells. Ex vivo HF studies showed that downregulation of EGLN1, EGLN3 and SERPINF1 promoted HF growth, postponed HF catagen transition, and prolonged the anagen stage, suggesting that these genes may be potentially utilized as therapeutic targets for AGA.
Conclusions: We characterized key transcriptome changes in male AGA HFs, and found that HIF-1 pathway-related genes (EGLN1, EGLN3) and Wnt pathway inhibitors (SERPINF1, SFRP2) may play important roles in AGA. What is already known about this topic? Multiple differentially expressed genes and signalling pathways have been found between hair follicles (HFs) in the balding area (frontal and vertex regions) and nonbalding area (occipital region) of individuals with androgenetic alopecia (AGA). A whole-transcriptome atlas of the vertex and occipital region is lacking. What does this study add? We identified a number of differentially expressed genes and pathways between balding vertex and nonbalding occipital AGA HFs by using whole-transcriptome analyses. We identified pathways not previously reported in AGA, such as the hypoxia-inducible factor (HIF)-1 signalling pathway. We verified that HIF-1 pathway-related genes (EGLN1, EGLN3) and Wnt pathway inhibitors (PEDF, SFRP2) played important roles in dermal papilla cell activity, hair growth and the hair cycle. What is the translational message? The EGLN1, EGLN3, SERPINF1 and SFRP2 genes may be potentially utilized as therapeutic targets for AGA.
© 2022 The Authors. British Journal of Dermatology published by John Wiley & Sons Ltd on behalf of British Association of Dermatologists.
Conflict of interest statement
The authors declare they have no conflicts of interest.
Figures



Comment in
-
Transcriptomic analysis identifies regulators of the Wnt signalling and hypoxia-inducible factor pathways as possible mediators of androgenetic alopecia.Br J Dermatol. 2022 Dec;187(6):845. doi: 10.1111/bjd.21881. Epub 2022 Oct 5. Br J Dermatol. 2022. PMID: 36199226 Free PMC article.
References
-
- Kelly Y, Blanco A, Tosti A. Androgenetic alopecia: an update of treatment options. Drugs 2016; 76:1349–64. - PubMed
-
- Ellis JA, Sinclair R, Harrap SB. Androgenetic alopecia: pathogenesis and potential for therapy. Expert Rev Mol Med 2002; 4:1–11. - PubMed
-
- Sawaya ME. Purification of androgen receptors in human sebocytes and hair. J Invest Dermatol 1992; 98 (6 Suppl.):92S–96S. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

