Historical contingencies and phage induction diversify bacterioplankton communities at the microscale
- PMID: 35862452
- PMCID: PMC9335236
- DOI: 10.1073/pnas.2117748119
Historical contingencies and phage induction diversify bacterioplankton communities at the microscale
Abstract
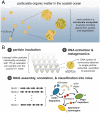
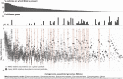
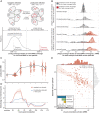
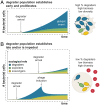
In many natural environments, microorganisms decompose microscale resource patches made of complex organic matter. The growth and collapse of populations on these resource patches unfold within spatial ranges of a few hundred micrometers or less, making such microscale ecosystems hotspots of heterotrophic metabolism. Despite the potential importance of patch-level dynamics for the large-scale functioning of heterotrophic microbial communities, we have not yet been able to delineate the ecological processes that control natural populations at the microscale. Here, we address this challenge by characterizing the natural marine communities that assembled on over 1,000 individual microscale particles of chitin, the most abundant marine polysaccharide. Using low-template shotgun metagenomics and imaging, we find significant variation in microscale community composition despite the similarity in initial species pools across replicates. Chitin-degrading taxa that were rare in seawater established large populations on a subset of particles, resulting in a wide range of predicted chitinolytic abilities and biomass at the level of individual particles. We show, through a mathematical model, that this variability can be attributed to stochastic colonization and historical contingencies affecting the tempo of growth on particles. We find evidence that one biological process leading to such noisy growth across particles is differential predation by temperate bacteriophages of chitin-degrading strains, the keystone members of the community. Thus, initial stochasticity in assembly states on individual particles, amplified through ecological interactions, may have significant consequences for the diversity and functionality of systems of microscale patches.
Keywords: community assembly; historical contingencies; marine particles; microscale; prophages.
Conflict of interest statement
The authors declare no competing interest.
Figures

References
-
- O’Donnell A. G., Young I. M., Rushton S. P., Shirley M. D., Crawford J. W., Visualization, modelling and prediction in soil microbiology. Nat. Rev. Microbiol. 5, 689–699 (2007). - PubMed
-
- McGlynn S. E., Chadwick G. L., Kempes C. P., Orphan V. J., Single cell activity reveals direct electron transfer in methanotrophic consortia. Nature 526, 531–535 (2015). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

