Potent universal beta-coronavirus therapeutic activity mediated by direct respiratory administration of a Spike S2 domain-specific human neutralizing monoclonal antibody
- PMID: 35862475
- PMCID: PMC9302814
- DOI: 10.1371/journal.ppat.1010691
Potent universal beta-coronavirus therapeutic activity mediated by direct respiratory administration of a Spike S2 domain-specific human neutralizing monoclonal antibody
Abstract
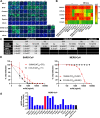
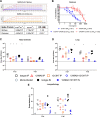
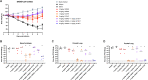
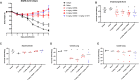
Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) marks the third novel β-coronavirus to cause significant human mortality in the last two decades. Although vaccines are available, too few have been administered worldwide to keep the virus in check and to prevent mutations leading to immune escape. To determine if antibodies could be identified with universal coronavirus activity, plasma from convalescent subjects was screened for IgG against a stabilized pre-fusion SARS-CoV-2 spike S2 domain, which is highly conserved between human β-coronavirus. From these subjects, several S2-specific human monoclonal antibodies (hmAbs) were developed that neutralized SARS-CoV-2 with recognition of all variants of concern (VoC) tested (Beta, Gamma, Delta, Epsilon, and Omicron). The hmAb 1249A8 emerged as the most potent and broad hmAb, able to recognize all human β-coronavirus and neutralize SARS-CoV and MERS-CoV. 1249A8 demonstrated significant prophylactic activity in K18 hACE2 mice infected with SARS-CoV-2 lineage A and lineage B Beta, and Omicron VoC. 1249A8 delivered as a single 4 mg/kg intranasal (i.n.) dose to hamsters 12 hours following infection with SARS-CoV-2 Delta protected them from weight loss, with therapeutic activity further enhanced when combined with 1213H7, an S1-specific neutralizing hmAb. As little as 2 mg/kg of 1249A8 i.n. dose 12 hours following infection with SARS-CoV Urbani strain, protected hamsters from weight loss and significantly reduced upper and lower respiratory viral burden. These results indicate in vivo cooperativity between S1 and S2 specific neutralizing hmAbs and that potent universal coronavirus neutralizing mAbs with therapeutic potential can be induced in humans and can guide universal coronavirus vaccine development.
Conflict of interest statement
I have read the journal’s policy and the authors of this manuscript have the following competing interests: M.S.P., J.-G.P., A.D., F.S.O., M.B., S.S., N.B.E., P.A.G., M.R.W., L.M.-S., and J.J.K. are co-inventors on patents that include claims related to the hmAbs described. A.L., D.C., J.W., P.L., and V.L.T. are employees of Aridis Pharmaceuticals.
Figures



Update of
-
Potent universal-coronavirus therapeutic activity mediated by direct respiratory administration of a Spike S2 domain-specific human neutralizing monoclonal antibody.bioRxiv [Preprint]. 2022 Mar 7:2022.03.05.483133. doi: 10.1101/2022.03.05.483133. bioRxiv. 2022. Update in: PLoS Pathog. 2022 Jul 21;18(7):e1010691. doi: 10.1371/journal.ppat.1010691. PMID: 35291292 Free PMC article. Updated. Preprint.
References
-
- Papanikolaou V, Chrysovergis A, Ragos V, Tsiambas E, Katsinis S, Manoli A, et al. From delta to Omicron: S1-RBD/S2 mutation/deletion equilibrium in SARS-CoV-2 defined variants. Gene. 2022;814:146134. Epub 20220104. doi: 10.1016/j.gene.2021.146134 ; PubMed Central PMCID: PMC8725615. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

