Erythroblastic islands foster granulopoiesis in parallel to terminal erythropoiesis
- PMID: 35862735
- PMCID: PMC9707396
- DOI: 10.1182/blood.2022015724
Erythroblastic islands foster granulopoiesis in parallel to terminal erythropoiesis
Abstract
The erythroblastic island (EBI), composed of a central macrophage surrounded by maturing erythroblasts, is the erythroid precursor niche. Despite numerous studies, its precise composition is still unclear. Using multispectral imaging flow cytometry, in vitro island reconstitution, and single-cell RNA sequencing of adult mouse bone marrow (BM) EBI-component cells enriched by gradient sedimentation, we present evidence that the CD11b+ cells present in the EBIs are neutrophil precursors specifically associated with BM EBI macrophages, indicating that erythro-(myelo)-blastic islands are a site for terminal granulopoiesis and erythropoiesis. We further demonstrate that the balance between these dominant and terminal differentiation programs is dynamically regulated within this BM niche by pathophysiological states that favor granulopoiesis during anemia of inflammation and favor erythropoiesis after erythropoietin stimulation. Finally, by molecular profiling, we reveal the heterogeneity of EBI macrophages by cellular indexing of transcriptome and epitope sequencing of mouse BM EBIs at baseline and after erythropoietin stimulation in vivo and provide a searchable online viewer of these data characterizing the macrophage subsets serving as hematopoietic niches. Taken together, our findings demonstrate that EBIs serve a dual role as niches for terminal erythropoiesis and granulopoiesis and the central macrophages adapt to optimize production of red blood cells or neutrophils.
© 2022 by The American Society of Hematology.
Figures
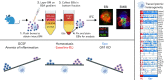
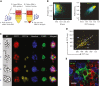
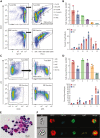

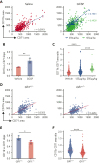


Comment in
-
Two's company but three's a blast!Blood. 2022 Oct 6;140(14):1578-1580. doi: 10.1182/blood.2022017644. Blood. 2022. PMID: 36201331 No abstract available.
References
-
- Bessis M. L’ilot erythroblastique. Unite functionelle de la moelle osseuse. Erythroblastic island, functional unity of bone marrowRev Hematol (Paris) 1958;13(1):8–11. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

