Metabolite Damage and Damage Control in a Minimal Genome
- PMID: 35862786
- PMCID: PMC9426524
- DOI: 10.1128/mbio.01630-22
Metabolite Damage and Damage Control in a Minimal Genome
Abstract
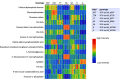
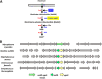
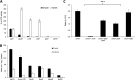
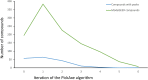
Analysis of the genes retained in the minimized Mycoplasma JCVI-Syn3A genome established that systems that repair or preempt metabolite damage are essential to life. Several genes known to have such functions were identified and experimentally validated, including 5-formyltetrahydrofolate cycloligase, coenzyme A (CoA) disulfide reductase, and certain hydrolases. Furthermore, we discovered that an enigmatic YqeK hydrolase domain fused to NadD has a novel proofreading function in NAD synthesis and could double as a MutT-like sanitizing enzyme for the nucleotide pool. Finally, we combined metabolomics and cheminformatics approaches to extend the core metabolic map of JCVI-Syn3A to include promiscuous enzymatic reactions and spontaneous side reactions. This extension revealed that several key metabolite damage control systems remain to be identified in JCVI-Syn3A, such as that for methylglyoxal. IMPORTANCE Metabolite damage and repair mechanisms are being increasingly recognized. We present here compelling genetic and biochemical evidence for the universal importance of these mechanisms by demonstrating that stripping a genome down to its barest essentials leaves metabolite damage control systems in place. Furthermore, our metabolomic and cheminformatic results point to the existence of a network of metabolite damage and damage control reactions that extends far beyond the corners of it that have been characterized so far. In sum, there can be little room left to doubt that metabolite damage and the systems that counter it are mainstream metabolic processes that cannot be separated from life itself.
Keywords: comparative genomics; hydrolase; metabolite repair; metabolomics; minimal genome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Hutchison CA, Chuang R-Y, Noskov VN, Assad-Garcia N, Deerinck TJ, Ellisman MH, Gill J, Kannan K, Karas BJ, Ma L, Pelletier JF, Qi Z-Q, Richter RA, Strychalski EA, Sun L, Suzuki Y, Tsvetanova B, Wise KS, Smith HO, Glass JI, Merryman C, Gibson DG, Venter JC. 2016. Design and synthesis of a minimal bacterial genome. Science 351:aad6253. doi: 10.1126/science.aad6253. - DOI - PubMed

