Comprehensive Evaluation of RNA and DNA Viromic Methods Based on Species Richness and Abundance Analyses Using Marmot Rectal Samples
- PMID: 35862817
- PMCID: PMC9426427
- DOI: 10.1128/msystems.00430-22
Comprehensive Evaluation of RNA and DNA Viromic Methods Based on Species Richness and Abundance Analyses Using Marmot Rectal Samples
Abstract
Viral metagenomics is the most powerful tool to profile viromic composition for a given sample. Different viromic methods, including amplification-free ones, have been developed, but choosing them for different purposes requires comprehensive benchmarks. Here, we assessed the performance of four routinely used methods, i.e., multiple displacement amplification (MDA), direct metagenomic sequencing (MTG), sequence-independent single-primer amplification (SIA), and metatranscriptomic sequencing (MTT), using marmot rectal samples as the templates spiked with five known viruses of different genome types. The obtained clean data were differently contaminated by host and bacterial genomes, resulting in MDA having the most, with ~72.1%, but MTT had only ~7.5% data, useful for follow-up viromic analysis. MDA showed a broader spectrum with higher efficiency to profile the DNA virome, and MTT captured almost all RNA viruses with extraordinary sensitivity; hence, they are advisable in richness-based viromic studies. MTG was weak in capturing single-stranded DNA viruses, and SIA could detect both RNA and DNA viruses but with high randomness. Due to biases to certain types of viruses, the four methods caused different alterations to species abundance compared to the initial virus composition. SIA and MDA introduced greater stochastic errors to relative abundances of species, genus, and family taxa, whereas the two amplification-free methods were more tolerant toward such errors and thus are recommendable in abundance-based analyses. In addition, genus taxon is a compromising analytic level that ensures technically supported and biologically and/or ecologically meaningful viromic conclusions. IMPORTANCE Viral metagenomics can be roughly divided into species richness-based studies and species abundance-based analyses. Viromic methods with different principles have been developed, but rational selection of these techniques according to different purposes requires comprehensive understanding of their properties. By assessing the four most widely used methods using template samples, we found that multiple displacement amplification (MDA) and metatranscriptomic sequencing (MTT) are advisable for species richness-based viromic studies, as they show excellent efficiency to detect DNA and RNA viruses. Meanwhile, metagenomic sequencing (MTG) and MTT are more compatible with stochastic errors of methods introduced into relative abundance of viromic taxa and hence are rational choices in species abundance-based analyses. This study also highlights that MTG needs to tackle host genome contamination and ameliorate the capacity to detect single-stranded DNA viruses in the future, and the MTT method requires an improvement in bacterial rRNA depletion prior to library preparation.
Keywords: performance comparison; species abundance; species richness; stochastic error; taxonomic rank; viral metagenomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




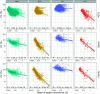
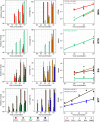
Similar articles
-
Phytovirome Analysis of Wild Plant Populations: Comparison of Double-Stranded RNA and Virion-Associated Nucleic Acid Metagenomic Approaches.J Virol. 2019 Dec 12;94(1):e01462-19. doi: 10.1128/JVI.01462-19. Print 2019 Dec 12. J Virol. 2019. PMID: 31597769 Free PMC article.
-
Assessment of REPLI-g Multiple Displacement Whole Genome Amplification (WGA) Techniques for Metagenomic Applications.J Biomol Tech. 2017 Apr;28(1):46-55. doi: 10.7171/jbt.17-2801-008. Epub 2017 Mar 21. J Biomol Tech. 2017. PMID: 28344519 Free PMC article.
-
Viral Metagenome-Based Precision Surveillance of Pig Population at Large Scale Reveals Viromic Signatures of Sample Types and Influence of Farming Management on Pig Virome.mSystems. 2021 Jun 29;6(3):e0042021. doi: 10.1128/mSystems.00420-21. Epub 2021 Jun 8. mSystems. 2021. PMID: 34100634 Free PMC article.
-
Emerging technologies in the study of the virome.Curr Opin Virol. 2022 Jun;54:101231. doi: 10.1016/j.coviro.2022.101231. Epub 2022 May 25. Curr Opin Virol. 2022. PMID: 35643020 Review.
-
Metagenomic characterization of viral communities in corals: mining biological signal from methodological noise.Environ Microbiol. 2015 Oct;17(10):3440-9. doi: 10.1111/1462-2920.12803. Epub 2015 Mar 27. Environ Microbiol. 2015. PMID: 25708646 Review.
Cited by
-
Detection and Characterization of a Reassortant Mammalian Orthoreovirus Isolated from Bats in Xinjiang, China.Viruses. 2022 Aug 27;14(9):1897. doi: 10.3390/v14091897. Viruses. 2022. PMID: 36146702 Free PMC article.
-
Unveiling bat-borne viruses: a comprehensive classification and analysis of virome evolution.Microbiome. 2024 Nov 14;12(1):235. doi: 10.1186/s40168-024-01955-1. Microbiome. 2024. PMID: 39543683 Free PMC article.
-
Virome Profiling of an Amur leopard cat Reveals Multiple Anelloviruses and a Bocaparvovirus.Vet Sci. 2022 Nov 17;9(11):640. doi: 10.3390/vetsci9110640. Vet Sci. 2022. PMID: 36423089 Free PMC article.
-
Isolation and Identification of Severe Fever with Thrombocytopenia Syndrome Virus from Farmed Mink in Shandong, China.Transbound Emerg Dis. 2024 Apr 5;2024:9604673. doi: 10.1155/2024/9604673. eCollection 2024. Transbound Emerg Dis. 2024. PMID: 40303144 Free PMC article.
-
Virome Profiling of Chickens with Hepatomegaly Rupture Syndrome Reveals Coinfection of Multiple Viruses.Viruses. 2023 May 26;15(6):1249. doi: 10.3390/v15061249. Viruses. 2023. PMID: 37376549 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
