Natural Competence in the Filamentous, Heterocystous Cyanobacterium Chlorogloeopsis fritschii PCC 6912
- PMID: 35862819
- PMCID: PMC9429965
- DOI: 10.1128/msphere.00997-21
Natural Competence in the Filamentous, Heterocystous Cyanobacterium Chlorogloeopsis fritschii PCC 6912
Abstract
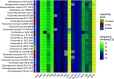
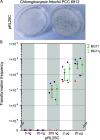
Lateral gene transfer plays an important role in the evolution of genetic diversity in prokaryotes. DNA transfer via natural transformation depends on the ability of recipient cells to actively transport DNA from the environment into the cytoplasm, termed natural competence, which relies on the presence of type IV pili and other competence proteins. Natural competence has been described in cyanobacteria for several organisms, including unicellular and filamentous species. However, natural competence in cyanobacteria that differentiate specialized cells for N2-fixation (heterocysts) and form branching or multiseriate cell filaments (termed subsection V) remains unknown. Here, we show that genes essential for natural competence are conserved in subsection V cyanobacteria. Furthermore, using the replicating plasmid pRL25C, we experimentally demonstrate natural competence in a subsection V organism: Chlorogloeopsis fritschii PCC 6912. Our results suggest that natural competence is a common trait in cyanobacteria forming complex cell filament morphologies. IMPORTANCE Cyanobacteria are crucial players in the global biogeochemical cycles, where they contribute to CO2- and N2-fixation. Their main ecological significance is the primary biomass production owing to oxygenic photosynthesis. Cyanobacteria are a diverse phylum, in which the most complex species differentiate specialized cell types and form true-branching or multiseriate cell filament structures (termed subsection V cyanobacteria). These bacteria are considered a peak in the evolution of prokaryotic multicellularity. Among others, species in that group inhabit fresh and marine water habitats, soil, and extreme habitats such as thermal springs. Here, we show that the core genes required for natural competence are frequent in subsection V cyanobacteria and demonstrate for the first time natural transformation in a member of subsection V. The prevalence of natural competence has implications for the role of DNA acquisition in the genome evolution of cyanobacteria. Furthermore, the presence of mechanisms for natural transformation opens up new possibilities for the genetic modification of subsection V cyanobacteria.
Keywords: cyanobacteria; genome evolution; lateral gene transfer; natural transformation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Grigorieva G, Shestakov S. 1982. Transformation in the cyanobacterium Synechocystis sp. 6803. FEMS Microbiol Lett 13:367–370. doi: 10.1111/j.1574-6968.1982.tb08289.x. - DOI
Publication types
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

